| Journal of Clinical Medicine Research, ISSN 1918-3003 print, 1918-3011 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Clin Med Res and Elmer Press Inc |
| Journal website https://www.jocmr.org |
Short Communication
Volume 15, Number 8-9, September 2023, pages 423-429
Computer-Aided Pulmonary Fibrosis Detection Leveraging an Advanced Artificial Intelligence Triage and Notification Software
Kavitha C. Selvana, d, Angad Kalrab, Joshua Reicherb, c, Michael Muellyb, c, Ayodeji Adegunsoyea
aSection of Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Chicago Medicine, Chicago, IL, USA
bIMVARIA Inc., Berkley, CA 94705, USA
cDepartment of Radiology, Stanford University, Stanford, CA, USA
dCorresponding Author: Kavitha C. Selvan, Section of Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Chicago Medicine, Chicago, IL 60637, USA
Manuscript submitted August 30, 2023, accepted September 25, 2023, published online September 30, 2023
Short title: Computer-Aided Pulmonary Fibrosis Detection
doi: https://doi.org/10.14740/jocmr5020
| Abstract | ▴Top |
Background: Improvement in recognition and referral of pulmonary fibrosis (PF) is vital to improving patient outcomes within interstitial lung disease. We determined the performance metrics and processing time of an artificial intelligence triage and notification software, ScreenDx-LungFibrosis™, developed to improve detection of PF.
Methods: ScreenDx-LungFibrosis™ was applied to chest computed tomography (CT) scans from multisource data. Device output (+/- PF) was compared to clinical diagnosis (+/- PF), and diagnostic performance was evaluated. Primary endpoints included device sensitivity and specificity > 80% and processing time < 4.5 min.
Results: Of 3,018 patients included, PF was present in 22.9%. ScreenDx-LungFibrosis™ detected PF with a sensitivity and specificity of 91.3% (95% confidence interval (CI): 89.0-93.3%) and 95.1% (95% CI: 94.2-96.0%), respectively. Mean processing time was 27.6 s (95% CI: 26.0 - 29.1 s).
Conclusions: ScreenDx-LungFibrosis™ accurately and reliably identified PF with a rapid per-case processing time, underscoring its potential for transformative improvement in PF outcomes when routinely applied to chest CTs.
Keywords: Pulmonary fibrosis; Interstitial lung disease; Early detection; Artificial intelligence
| Introduction | ▴Top |
Interstitial lung disease (ILD) encompasses a spectrum of heterogenous parenchymal disorders, many of which are characterized by the development and progression of pulmonary fibrosis (PF). The presence of PF confers poor clinical outcomes, which may in part be due to a delay in disease recognition and subsequent referral to a tertiary care center. The factors underpinning these delays are multifactorial, due in part to frequent misdiagnosis with more commonly encountered diseases such as asthma or emphysema and underreporting of radiographic features characteristic of ILD [1-4]. Establishing concrete methods to increase recognition and referral of early PF is vital to improving patient outcomes.
In recent years, there has been rapid expansion in the use of artificial intelligence (AI) algorithms to classify and predict outcomes in ILD [5-8]. However, few studies have focused on the use of AI for screening and triage of PF. We hypothesize that a highly accurate and precise triage tool that requires minimal per-case processing time could be leveraged to improve outcomes within ILD. The objective of this study was to externally validate the performance metrics and processing time of an AI software, ScreenDx-LungFibrosis™, developed to improve detection of PF [9].
| Materials and Methods | ▴Top |
Study data
This study leveraged public and private clinical trial and registry data provided by the National Institutes of Health (NIH), The Cancer Imaging Archive (TCIA), and Open Source Imaging Consortium (OSIC). Patient demographics, chest computed tomography (CT) scans, and site-specific diagnoses were collected. We consulted extensively with the Argus Institutional Review Board (IRB) who determined that our study did not need ethical approval. An IRB official waiver of ethical approval was granted from the Argus IRB (adjudication #081920). The data protocols are in accordance with the ethical standards of our institution and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Study population
We performed a retrospective analysis of patients ≥ 22 years old (per the Food and Drug Administration (FDA) Center for Devices and Radiologic Health definition of adulthood [10]) with available chest CTs and site-specific PF diagnoses (categorized as present or absent). PF was defined broadly as any fibrotic ILD (fILD; physician confirmed diagnosis of ILD with fibrotic features, including reticulation, honeycombing, traction bronchiectasis, and/or architectural distortion). The diagnosis of PF was made by physician determination at the respective sites and supported by the International Classification of Diseases: Ninth Revision (ICD-9) diagnosis codes where available within the electronic medical record. The majority of PF diagnoses were achieved via gold-standard multidisciplinary discussion (MDD) (Table 1).
 Click to view | Table 1. Baseline Cohort Characteristics |
CT parameters
ScreenDx-LungFibrosis™ was designed to detect PF across a broad range of CT specifications, therefore, a variety of thin-acquisition and axial-reconstruction images were obtained. CT image slice thickness were ≤ 5 mm and included a full view of both lungs, with minimal motion artifact, imaging noise, blooming, or misregistration. The sharpest available reconstruction kernel per study was used for analysis. No images used during device training were utilized in this protocol.
Model design
ScreenDx-LungFibrosis™ is a deep learning, convolutional neural network designed to identify CT imaging features consistent with PF (Fig. 1). It was developed as an adjunct for flagging and triaging cases suspicious for PF. The algorithm was trained on a multisource international dataset of > 3,600 patients and finetuned within a USA-based pulmonary cohort of 381 patients. Data were split into training, tuning, and “holdout” testing sets that were used to ensure generalizability of the model. In development and testing, the model area under the receiver operating characteristic (AUROC) was 0.997 (95% confidence interval (CI): 0.98 - 1.00), with a sensitivity and specificity of 100% and 98%, respectively [9].
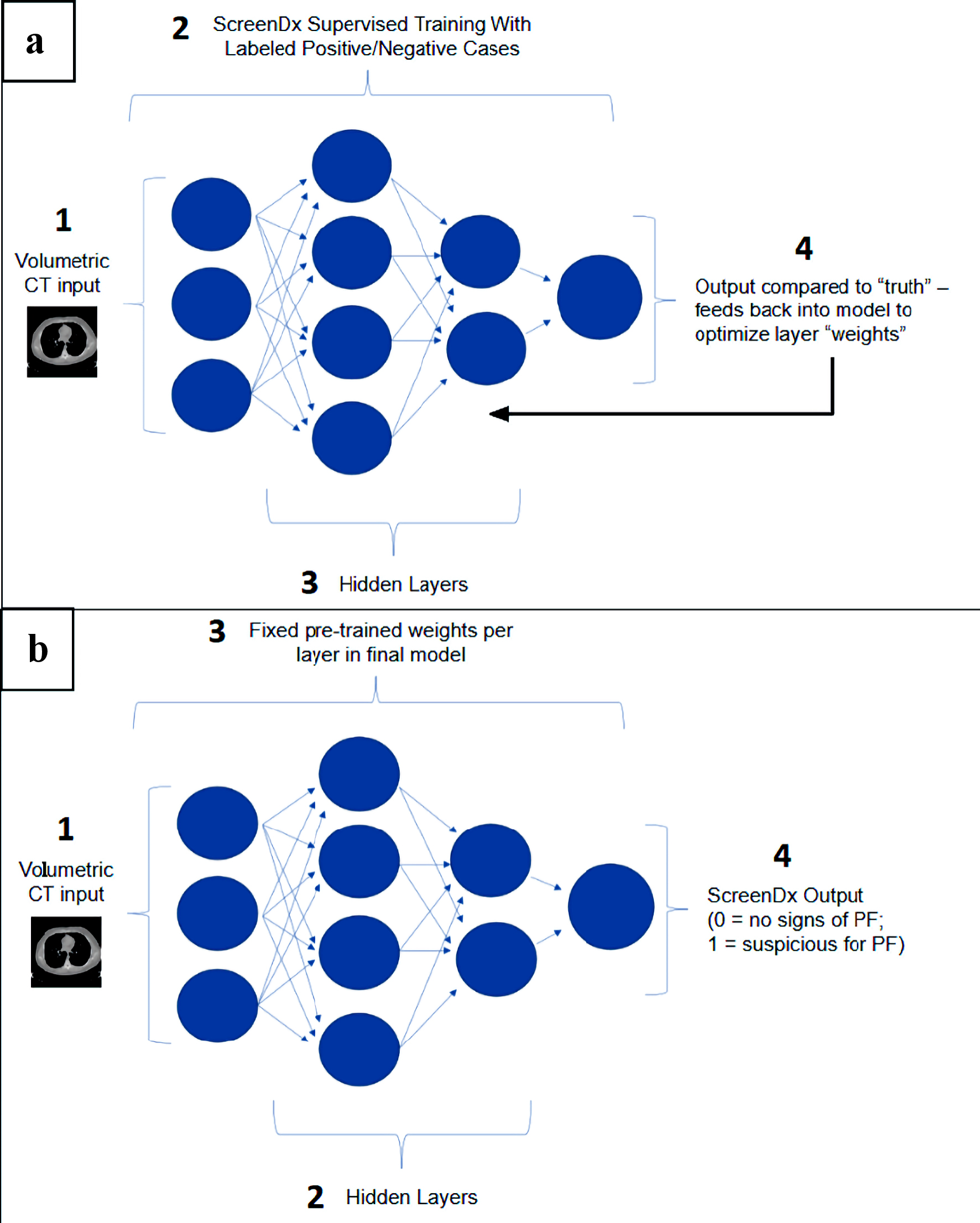 Click for large image | Figure 1. ScreenDx-LungFibrosis™ analysis algorithm. (a) ScreenDx-LungFibrosis™ supervised training: The analysis algorithm receives volumetric CT input normalized to standardized pixel values (1) from thousands of labeled cases (i.e., positive or negative for PF). It is then trained (2) using a Convolutional Neural Network (CNN) that incorporates thousands of features via hidden layers (3) into the network. Algorithm output (4) is compared to the “truth”, established by the labeled cases, and feeds back into the model to optimize layer weights. (b) ScreenDx-LungFibrosis™ evaluation of new cases. The analysis algorithm receives volumetric CT input normalized to standardized pixel values (1) from new cases, which proceed through pre-trained hidden layers (2) with fixed weights per layer (3) to create a binary output (4) of 0 (no signs of PF) or 1 (suspicious for PF). PF: pulmonary fibrosis; CT: computed tomography. |
Study outcomes
Primary study endpoints were: 1) model sensitivity and specificity > 80% compared to site-specific diagnosis; and 2) case processing time to notification generation < 4.5 min (95% CI: 4.1 - 4.8), evaluated in a randomly selected subset of 300 patients. The sensitivity/specificity and processing time thresholds were chosen based on FDA performance cutoffs for computer-aided detection triage tools (e.g., K201020, DEN170073, K182875) [11].
Statistical analysis
Sensitivity and specificity outcomes were compared to the 80% targets based on one-sided hypothesis tests examining the lower 95% confidence bound (97.5% level). Baseline patient and CT characteristics were summarized using descriptive statistics. Model output was analyzed using AUROC, and performance metrics were determined. All analyses were conducted using Stata software (17.1).
| Results | ▴Top |
Cohort characteristics
Of the 13,354 patients evaluated, 3,018 met inclusion criteria (Supplementary Material 1, www.jocmr.org). The median age was 65 years, and 55.5% (n = 1,678) were male (Table 1). Patients with PF were significantly older than those without PF (69.9 years vs. 63.2 years, P < 0.001). The majority of patients were ever-smokers (61.8%, n = 1,865), and of those with available race/ethnicity data (n = 1,662), 84.9% (n = 1,411) were White.
PF was present in 22.9% (n = 692) of patients. Among subjects with PF, idiopathic pulmonary fibrosis (IPF) was most common (n = 562, 81.2%). In patients without PF, 35.5% had normal CT imaging without evidence of disease (n = 1,072), 14.2% had cancer (n = 429), and 12.3% had coronavirus disease 2019 (COVID-19) (n = 371). The average CT slice thickness was 1.6 mm in patients with PF, compared to 2.4 mm in those without PF (P < 0.001). CT manufacturers are listed here (Supplementary Material 2, www.jocmr.org).
Device performance
ScreenDx-LungFibrosis™ had a sensitivity and specificity for detecting PF of 91.3% (95% CI: 89.0-93.3%) and 95.1% (95% CI: 94.2-96.0%), respectively, which met the prespecified primary endpoints (Table 2). The device performed well under conditions with both low (10%) and high (50%) prevalence of PF (positive predictive value (PPV) of 66.9% and 94.8%, and a negative predictive value (NPV) of 99.0% and 91.4%, respectively). Patients that screened positive for PF were 19 times more likely to have PF compared to those that screened negative (likelihood ratio (LR)+ 18.8, 95% CI: 15.8 - 22.9), and the odds of detecting PF in the study population were 206.3 (95% CI: 149 - 286). The mean processing time was 10-fold faster than the pre-specified endpoint (27.6 s, 95% CI: 26.0 - 29.1 s vs. 4.5 min, 95% CI: 4.1 - 4.8 min; P < 0.0001).
 Click to view | Table 2. Diagnostic Parameters of ScreenDx-LungFibrosis™ |
| Discussion | ▴Top |
In this large external validation study leveraging multisource, international data, we demonstrate that ScreenDx-LungFibrosis™ accurately and reliably identified PF with a strikingly rapid per-case processing time. As delays in recognition and timely referral of PF contribute significantly to morbidity and mortality within ILD, we believe that routine application of this screening and triage tool, particularly within the context of studies ordered in the emergency department (ED) or for high-risk populations, would lead to improved patient outcomes.
Due to a variety of factors, patients with ILD often present to care late in the course of disease, at which time, significant and irreversible PF has developed, and treatment is limited. The ED is one such setting that likely contributes to these delays. Here, clinicians focus on obtaining preliminary imaging interpretations that answer a specific and emergent question. Within that context, complete interpretations that include critical incidental findings such as PF may not be provided or communicated to patients until after discharge. Additionally, patients at high risk for developing PF, such as those with a tobacco use history [12], or prior COVID-19 infection [13, 14], may have evidence of subtle or early PF on imaging scans obtained for alternative purposes (e.g., lung cancer screening), and could benefit from highly sensitive, computer-aided tools that augment radiology review. ScreenDx-LungFibrosis™ performs with a rapid per-case processing time and can be utilized across a broad range of CT manufacturers, highlighting its clear utility in improving early recognition and appropriate triage of PF in these settings.
Few other studies have focused on automated CT detection of ILD. High attenuation areas [15], interstitial lung abnormalities [7], and various features identified by densitometric and histogram-based measurements [16, 17] and texture analysis [18, 19] have all been evaluated as potential indicators of ILD and outcomes; however, these studies are suboptimal in providing a sensitive and specific method to objectively detect features of fibrosis, which confer the highest risk for progression and mortality. ScreenDx-LungFibrosis™ operates at a sensitivity and specificity > 90% for detecting PF, and if used routinely, could enable clinicians to identify patients at the highest risk for poor outcomes from ILD. Additionally, through consistent recognition and automated reporting of PF, this tool could be an asset to centers lacking subspecialized radiologists, thus enhancing their ability to recognize and report PF early. Our system is validated across a wide range of demographics, and while statistically, males with a tobacco history are most likely to be diagnosed with PF, the benefit of identification and early treatment in the broader population (e.g., females under 60 years old) have potential to be even more meaningful.
This study has several limitations: 1) Its retrospective study design using registry data that did not consistently include details on patient or site demographics; 2) The stage of PF was not consistently known, which limited the ability to evaluate tool performance in milder cases of PF; 3) As occurs with the use of any deep learning algorithm, the exact parameters evaluated by ScreenDx-LungFibrosis™ are unknown; and 4) Impact of this tool on clinical workflow and subspecialty pulmonary referral was not evaluated. Moving forward, piloting this technology to determine the feasibility of implementation is of critical importance.
In conclusion, ScreenDx-LungFibrosis™ accurately and reliably identified PF with a rapid per-case processing time, underscoring its potential for transformative improvement in outcomes when routinely applied in patients with fibrotic ILDs.
| Supplementary Material | ▴Top |
Suppl 1. Study flow diagram.
Suppl 2. Computed tomography manufacturers.
Acknowledgments
Thanks to Drs. Julia Seaman, PhD and Isabel Allen, PhD for biostatistical analysis, to Scott Gellert for guidance on the clinical study protocol, and to FGCL-3019-049 Trial, PRAISE Trial, Harvard COVID-19 Dataset, the National Institutes of Health (NIH), Medical Imaging and Data Resource Center (MIDRC), National Lung Cancer Screening Trial (NLST), Open Source Imaging Consortium (OSIC), and Babak-Tehran Dataset for data acquisition.
Financial Disclosure
Dr. Selvan is supported by the National Institutes of Health (NIH)/National Heart, Lung, and Blood (NHLBI) grant T32HL007605. Dr. Adegunsoye is supported by the NIH grant K23HL146942. Foundations: CHEST Foundation, and Pulmonary Fibrosis Foundation.
Conflict of Interest
Dr. Reicher, Dr. Muelly, and Mr. Scott Gellert have a financial interest in IMVARIA Inc. Dr. Adegunsoye has relationships with Genentech Inc., Inogen Inc., Medscape LLC, PatientMpower, Abbvie, and Boehringer Ingelheim Corp USA.
Informed Consent
Informed consent was obtained from all participants included in this study.
Author Contributions
Kavitha C. Selvan wrote the main manuscript text, contributed to data analysis, and prepared Tables 1, 2. Angad Kalra led the study of methodology and software design. Joshua Reicher led study conceptualization, contributed to data analysis, and prepared Figure 1. Michael Muelly. led project administration and contributed to project resources. Ayodeji Adegunsoye contributed to study methodology and led manuscript supervision. All authors read and reviewed the previous versions of the manuscript and the final manuscript.
Data Availability
Any inquiries regarding supporting data availability of this study should be directed to the corresponding author.
Abbreviations
AI: artificial intelligence; AUROC: area under the receiver operating characteristic; CT: computed tomography; ED: emergency department; FDA: Food and Drug Administration; ICD: International Classification of Diseases; IPF: idiopathic pulmonary fibrosis; ILD: interstitial lung disease; NPV: negative predictive value; PF: pulmonary fibrosis; PPV: positive predictive value
| References | ▴Top |
- Rahman KKM, Samaria JK. Diagnostic delay and misdiagnosis in interstitial lung disease at primary health care level. Eur Respir J. 2016;48(60):PA861.
- Pritchard D, Adegunsoye A, Lafond E, Pugashetti JV, DiGeronimo R, Boctor N, Sarma N, et al. Diagnostic test interpretation and referral delay in patients with interstitial lung disease. Respir Res. 2019;20(1):253.
doi pubmed pmc - Mooney J, Chang E, Lalla D, Papoyan E, Raimundo K, Reddy SR, Stauffer J, et al. Potential Delays in Diagnosis of Idiopathic Pulmonary Fibrosis in Medicare Beneficiaries. Ann Am Thorac Soc. 2019;16(3):393-396.
doi pubmed pmc - Cosgrove GP, Bianchi P, Danese S, Lederer DJ. Barriers to timely diagnosis of interstitial lung disease in the real world: the INTENSITY survey. BMC Pulm Med. 2018;18(1):9.
doi pubmed pmc - Walsh SLF, Calandriello L, Silva M, Sverzellati N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med. 2018;6(11):837-845.
doi pubmed - Bermejo-Pelaez D, Ash SY, Washko GR, San Jose Estepar R, Ledesma-Carbayo MJ. Classification of interstitial lung abnormality patterns with an ensemble of deep convolutional neural networks. Sci Rep. 2020;10(1):338.
doi pubmed pmc - Walsh SLF, Mackintosh JA, Calandriello L, Silva M, Sverzellati N, Larici AR, Humphries SM, et al. Deep learning-based outcome prediction in progressive fibrotic lung disease using high-resolution computed tomography. Am J Respir Crit Care Med. 2022;206(7):883-891.
doi pubmed - Mei X, Liu Z, Singh A, Lange M, Boddu P, Gong JQX, Lee J, et al. Interstitial lung disease diagnosis and prognosis using an AI system integrating longitudinal data. Nat Commun. 2023;14(1):2272.
doi pubmed pmc - Touloumes N, Gagianas G, Bradley J, Kalra A, Muelly M, Reicher J. Visual Dx: An artificial intelligence-based algorithm for the incidental detection of pulmonary fibrosis. In submission.
- U.S. Food and Drug Administration. About the FDA’s Center for Devices and Radiological Health. FDA. 2023. https://www.fda.gov/about-fda/fda-organization/center-devices-and-radiological-health.
- Center for Devices and Radiological Health. Artificial Intelligence and Machine Learning (AI/Ml)-Enabled Medical D. U.S. Food and Drug Administration, Oct 5, 2022. www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices.
- Pezzuto A, Tammaro A, Salerno G, Borro M, Carico E. Lung nodule and pulmonary fibrosis at first evaluation in a smoker undergoing a smoking cessation program. Global J Respir Care. 2022;8:11-13.
- Cattaruzza MS, Gorini G, Bosetti C, Boffi R, Lugo A, Veronese C, Carreras G, et al. Covid-19 and the role of smoking: the protocol of the multicentric prospective study COSMO-IT (COvid19 and SMOking in ITaly). Acta Biomed. 2020;91(3):e2020062.
doi pubmed pmc - Adegunsoye A, Baccile R, Best TJ, Zaksas V, Zhang H, Karnik R, Patel BK, et al. Pharmacotherapy and pulmonary fibrosis risk after SARS-CoV-2 infection: a prospective nationwide cohort study in the United States. Lancet Reg Health Am. 2023;25:100566.
doi pubmed pmc - Podolanczuk AJ, Oelsner EC, Barr RG, Hoffman EA, Armstrong HF, Austin JH, Basner RC, et al. High attenuation areas on chest computed tomography in community-dwelling adults: the MESA study. Eur Respir J. 2016;48(5):1442-1452.
doi pubmed pmc - Matsuoka S, Yamashiro T, Matsushita S, Kotoku A, Fujikawa A, Yagihashi K, Nakajima Y. Quantitative CT evaluation in patients with combined pulmonary fibrosis and emphysema: correlation with pulmonary function. Acad Radiol. 2015;22(5):626-631.
doi pubmed - Ash SY, Harmouche R, Vallejo DL, Villalba JA, Ostridge K, Gunville R, Come CE, et al. Densitometric and local histogram based analysis of computed tomography images in patients with idiopathic pulmonary fibrosis. Respir Res. 2017;18(1):45.
doi pubmed pmc - Wang J, Li F, Doi K, Li Q. Computerized detection of diffuse lung disease in MDCT: the usefulness of statistical texture features. Phys Med Biol. 2009;54(22):6881-6899.
doi pubmed - Depeursinge A, Chin AS, Leung AN, Terrone D, Bristow M, Rosen G, Rubin DL. Automated classification of usual interstitial pneumonia using regional volumetric texture analysis in high-resolution computed tomography. Invest Radiol. 2015;50(4):261-267.
doi pubmed pmc
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Clinical Medicine Research is published by Elmer Press Inc.


