| Journal of Clinical Medicine Research, ISSN 1918-3003 print, 1918-3011 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Clin Med Res and Elmer Press Inc |
| Journal website http://www.jocmr.org |
Original Article
Volume 12, Number 6, June 2020, pages 369-376
Purifying Selection in Human Immunodeficiency Virus-1 pol Gene in Perinatally Human Immunodeficiency Virus-1-Infected Children Harboring Discordant Immunological Response and Virological Nonresponse to Long-Term Antiretroviral Therapy
Ralph-Sydney Mboumba Bouassaa, b, Helene Perea, c, Christian Diamant Mossoro-Kpinded, e, Pierre Roquesf, Jean Chrysostome Godyd, g, Sandrine Moussah, David Veyera, Gerard Gresenguetd, i, Charlotte Charpentierj, Mohammad-Ali Jenabiank, Joel Fleury Djoba Siawayab, l, Laurent Beleca, b, c, m
aLaboratoire de Virologie, Hopital Europeen Georges Pompidou, Assistance Publique-Hopitaux de Paris (AP-HP) and Universite de Paris, Paris Sorbonne Cite, Paris, France
bEcole Doctorale Regionale en Infectiologie Tropicale, Franceville, Gabon
cUniversite de Paris, Paris Sorbonne Cite, Paris, France
dFaculte des Sciences de la Sante, Universite de Bangui, Bangui, Central African Republic
eLaboratoire National de Biologie Clinique et de Sante Publique, Bangui, Central African Republic
fCommissariat a l’Energie Atomique (CEA)-Universite Paris-Saclay; INSERM U1184, Immunology of Viral Infections and Autoimmune Diseases (IMVA), IDMIT Department, Institut de Biologie Francois-Jacob (IBJF), Fontenay-aux-Roses, France
gComplexe Pediatrique, Bangui, Central African Republic
hInstitut Pasteur de Bangui, Bangui, Central African Republic
iUnite de Recherches et d’Intervention sur les Maladies Sexuellement Transmissibles et le SIDA, Departement de Sante Publique, Faculte des Sciences de la Sante de Bangui, Central African Republic
jIAME, UMR 1137, INSERM, Universite Paris Diderot, Sorbonne Paris Cite, AP-HP, Laboratoire de Virologie, Hopital Bichat, AP-HP, Paris, France
kDepartement des Sciences Biologiques et Centre de Recherche BioMed, Universite du Quebec a Montreal (UQAM), Montreal, QC, Canada
lLaboratory Medicine, Mother and Child University Hospital Jeanne Ebori, Libreville, Gabon
mCorresponding Author: Belec Laurent, Hopital Europeen Georges Pompidou, Laboratoire de Virologie, 20 rue Leblanc, 75015 Paris, France
Manuscript submitted April 7, 2020, accepted April 23, 2020, published online June 4, 2020
Short title: Genetic Selection of HIV-1 in Children
doi: https://doi.org/10.14740/jocmr4157
| Abstract | ▴Top |
Background: Biological monitoring of antiretroviral treatment (ART) in human immunodeficiency virus (HIV)-infected pediatric population remains challenging. The aim of the present study was to assess the long-term HIV-1 genetic diversity in pol gene in HIV-1-infected children in virological failure under antiretroviral regimen adapted according to the successive World Health Organization (WHO) guidelines for resource-constrained settings.
Methods: HIV-1 diversity in pol gene was assessed in HIV-1-infected children and adolescents born from HIV-infected mothers (median age at follow-up: 13.8 years) in virological failure (VF+) despite long-term regimen recommended by the WHO. The numbers of nonsynonymous substitutions per potential nonsynonymous site (dN) and of synonymous substitutions at potential synonymous sites (dS) in HIV-1 pol gene and the dN/dS ratios were used to estimate the selective pressure on circulating HIV-1.
Results: The immunological responses to ART basically corresponded to: 1) Full therapeutic failure with immunological (I-) and virological nonresponses in one-quarter (24.6%) of study children ((I-, VF+) subgroup); 2) Discordant immunovirological responses with paradoxical high CD4 T cell counts (I+) and high HIV-1 RNA load in the remaining cohort patients (75.4%) ((I+, VF+) subgroup). The mean dS was 1.8-fold higher in (I+, VF+) than (I-, VF+) subgroup (25.9 ± 18.4 vs. 14.3 ± 10.8). In the (I+, VF+) subgroup, the mean dS was 1.6-fold higher than the mean dN. Finally, the mean dN/dS ratio was 2.1-fold lower in (I+, VF+) than (I-, VF+) subgroup (0.6 ± 0.3 vs. 1.3 ± 0.7), indicating purifying selection in the immunovirological discordant (I+, VF+) subgroup and positive selection in the immunovirological failure (I-, VF+) subgroup.
Conclusions: Children and adolescents in immunovirological therapeutic failure harbor positive selection of HIV-1 strains favoring diversifying in pol-encoded amino acids. In contrast, children with persistent discordant immunovirological responses show accumulation of mutations and purifying selection in pol gene sequences, indicating limited genetic evolution and likely suggesting genetic adaptation of viruses to host functional constraints.
Keywords: Children; Adolescents; Virological failure; Immunovirological dissociation; Synonymous mutations; Nonsynonymous mutations; Purifying selection; HIV-1 pol gene
| Introduction | ▴Top |
Nearly 90% of human immunodeficiency virus (HIV)-infected children reside in sub-Saharan Africa [1]. Treating HIV-infected children in resource-limited settings remains tricky. Antiretroviral treatment (ART) in African pediatric populations remains associated with high rates of treatment failure ranging from 19% to 53% [2].
HIV-1 RNA load and CD4 T cell count constitute complementary surrogate biological markers to monitor ART in pediatric acquired immunodeficiency syndrome (AIDS) [3-5]. According to the World Health Organization (WHO) recommendations, the threshold for virological failure (VF+) in HIV-infected children and adolescents treated by ART was set at 1,000 copies/mL [4, 6-8]. In practice, the correlation between immunological and virological failures in pediatric cohorts appeared often minimal with frequent immunological and virological discordant responses during prolonged use of antiretroviral drugs [5, 9-11]. Beyond the classical quantitative surrogate markers, including HIV-1 RNA load and CD4 T cell count, the understanding of genetic evolution of circulating HIV-1 strains in HIV-infected children taking long-term ART and in virological failure could provide relevant insights to improve and adapt the management of ART-experienced children exhibiting immunological and virological discordant responses.
An observational cohort of HIV-infected children was prospectively established since 2007 at the Complexe Pediatrique of Bangui, the main health care clinic for HIV-infected children of the Central African Republic [12-14]. The aim of the present study was to assess the long-term HIV-1 genetic diversity in pol gene in HIV-1-infected children in virological failure under antiretroviral regimen adapted according to the successive WHO guidelines for resource-constrained settings [3, 4, 6].
| Materials and Methods | ▴Top |
Study population
A total of 220 HIV-1-infected children and adolescents born from HIV-1-infected mothers attending the Complexe Pediatrique of Bangui were prospectively recruited in 2009 and further followed up until 2013 for 39 months in a descriptive observational cohort study, assessing their outcome under ART, as previously reported elsewhere [13, 14]. The median age of the children was 13.8 years (interquartile range (IQR): 8 - 17), and the sex ratio (male/female) was 0.85 (101/119). The majority of the children and adolescents (n = 198, 90%) received first-line regimen according to the revised WHO recommendations of 2013 [4], for a mean duration in first-line of 65.6 months (range: 18.8 - 69.9). The remaining children and adolescents (n = 22, 10%) received second-line regimen for a mean duration of 78.2 months (range: 13.3 - 88.3). The most prescribed treatment was stavudine (d4T)/lamivudine (3TC)/nevirapine (NVP) combination. Only a minority (12.7% (28/220)) of children was under protease inhibitor (PI)-based regimen.
Virological and immunological categorization of children under ART
Plasma HIV-1 RNA load and CD4 T cell measurements were carried out, as previously described [14]. In brief, plasma HIV-1 RNA loads were measured using the Amplix® platform developed by Biosynex (Strasbourg, France), which integrates a fully automated station for nucleic acid extraction with a real-time polymerase chain reaction (PCR) amplification station, using lyophilized Amplix HIV-1 RNA quantitative reagents (Biosynex). The assay detects HIV-1 groups M and O and several circulating recombinant forms (CRFs). The CD4 T lymphocyte count was performed using the Apogee Auto40 flow cytometer from Apogee Flow Systems laboratories (Hemel Hempstead, London, UK).
Successful immunological response to therapy (I+) was defined as increase in CD4 T cells count according to the age at follow-up visit (i.e., CD4 T cells count > 750/µL for children less than 5 years and CD4 T cells count > 500/µL for children and adolescents older than 5 years) [3, 4].
Finally, children under ART in virological failure (VF+) according to the WHO threshold (HIV-1 RNA load > 1,000 copies/mL) were classified into two categories according to their immunovirological responses: (I+, VF+) for immunological responders but virological nonresponders; and (I-, VF+) for both immunological and virological nonresponders.
Detection of drug resistance mutations
Detection of drug resistance mutations (DRMs) was carried out in plasmas from randomly selected children with HIV-1 RNA load above 1,000 copies/mL after 39 months of follow-up, as reported [14]. Randomization for DRMs genotyping consisted in the selection of one out of three children from the (I+, VF+) subgroup and one out of two children in the (I-, VF+) subgroup.
Overtime genetic variability of HIV-1 RNA pol gene sequences
To evaluate whether, and to what extent, the genetic variability correlates with positive selection for amino acid changes, the accumulation rate of synonymous (S) and nonsynonymous (N) substitutions per potential synonymous and nonsynonymous site was analyzed in each patients by pairwise comparisons of HIV-1 sequences between inclusion and follow-up. The genetic variability overtime in the two subgroups of virological failure (I+, VF+) and (I-, VF+) was estimated using aligned sequences according to open-reading frame and codons by pairwise comparisons of all HIV-1 pol gene sequences obtained at baseline and after 39 months of prospective follow-up from selected children and adolescents of each subgroup, corresponding to the appraisal of 433 paired nucleotide positions. Analyses were conducted according to the Nei-Gojobori model [15], using the Jukes-Cantor correction for multiple substitutions implemented in the Molecular Evolutionary Genetics Analysis version 6 (MEGA6) program package (www.meagasoftware.net), as described [16]. All ambiguous positions or gap were removed for each sequence pair. For each patient, synonymous (dS) and nonsynonymous (dN) nucleotide differences between each pair of HIV-1 pol gene sequences at baseline and follow-up, corresponding to the genetic distance (d) between paired sequences, were determined, and the nonsynonymous/synonymous ratio rates (ω = dN/dS) were further calculated. The ω ratios are important indicators of selective pressure at the protein level, with ω = 1 meaning neutral mutations, ω < 1.0 purifying selection and ω > 1.0 diversifying positive selection favoring diversity at the amino acid level [17-19].
After validation, all nucleotide sequences were submitted to the European Nucleotide Archive at GenBankEMBL-EBI, with the following accession numbers: LT577626-LT577673; LT578170-LT578280 and LT726745-LT726792.
Statistical analyses
Means and standard deviations (SD) were calculated for quantitative variables using the software R (version 3.5.2). Average genetic distances were compared within each subgroup by the nonparametric Wilcoxon’s test for paired data, and between subgroups using the Welch’s t-test assuming unequal variances between subgroups.
| Ethics statement | ▴Top |
The study was formally approved by the Scientific Committee of Faculte des Sciences de la Sante (“FACSS”) of Bangui, Central African Republic, and was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
| Results | ▴Top |
Longitudinal immunovirological outcomes of the cohort children after 39 months of follow-up
The Figure 1 depicts the distribution of study children at inclusion and after the follow-up period, according to their immunological and virological responses to ART. At baseline in 2009, 130 (55.1%) children and adolescents among the 245 cohort children were in virological failure (VF+). Among them, 32 (24.6%) were immunological and virological nonresponders (I-, VF+), with a very low mean CD4 T cells count (mean: 226 ± 153 cells/µL) and high viral load (mean: 4.7 ± 1.7 log copies/mL), while the majority (n = 98; 75.4%) were immunological responders but virological nonresponders (I+, VF+) with normal CD4 T cell count (mean: 809 ± 280 cells/µL) and elevated viral load (mean: 4.8 ± 1.5 log copies/mL).
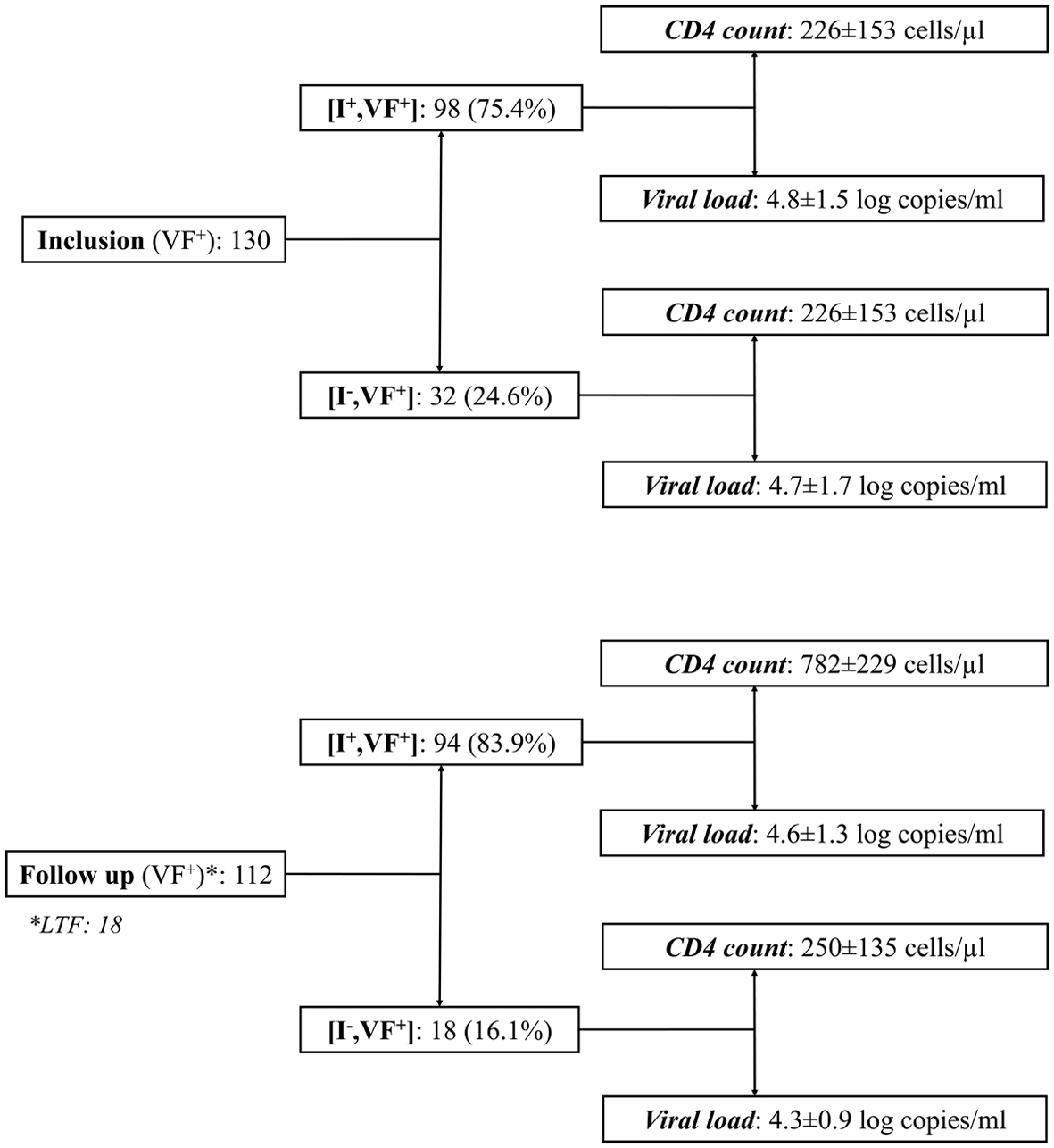 Click for large image | Figure 1. Distribution of study children in virological failure (VF+) at inclusion and after the follow-up period, according to their immunological (CD4 T cell count) and virological (circulating viral load) responses to antiretroviral treatment. Results are expressed as mean ± standard deviation. I+: immunological responders; I-: immunological nonresponders; LTF: lost-to-follow up; CD4: cluster of differentiation 4. |
After 39 months, only 18 (8.2%) children were lost to followed up, including three (1.3%) in the discordant (I+, VF+) subgroup and 15 (6.8%) in the immunovirological nonresponder subgroup. A total of 112 children and adolescents were in virological failure (VF+), including 18 (16.1%) immunological and virological nonresponders (I-, VF+), with low CD4 T cells count (mean: 250 ± 135 cells/µL) and high viral load (mean: 4.3 ± 0.9 log copies/mL) and 94 (83.9%) immunological responders but virological nonresponders (I+, FV+), with high CD4 T cell count (mean: 782 ± 229 cells/µL) and viral load (mean: 4.6 ± 1.3 log copies/mL).
Temporal variation of genetic diversity in HIV-1 RNA pol gene from circulating HIV-1 strains according to immunovirological subgroups of virological failure
Paired genotyping for DRMs in HIV-1 pol gene at baseline and after follow-up was obtained in viruses from 32 selected children in the (I+, VF+) subgroup and from nine selected children in the (I-, VF+) subgroup.
After 39 months of follow-up, 40 (97.5%) (including 31/32 (96.8%) in (I+, VF+) subgroup and 9/9 (100%) in (I-, VF+) subgroup) of the 41 genotypes from the children cohort in virological failure harbored genotype profiles of resistance to WHO-recommended first-line therapeutic regimen with a high frequency of natural polymorphisms in the protease gene sequences in over 90% of sequenced viruses (not shown). The mutation V82A, identified as being more frequent in patients with a good immunological response despite virological failure [20] was observed in viruses from five children belonging to the subgroup (I+, VF+). Most sequenced viruses remained susceptible to the majority of the WHO-recommended antiretroviral drugs (not shown).
Variation in genetic variability overtime between baseline and follow-up involved 64 HIV-1 pol gene nucleotide sequences in the (I+, VF+) subgroup and 18 in the (I-, VF+) subgroup (Fig. 2). The number of nonsynonymous substitutions per potential nonsynonymous site (dN), the number of synonymous substitutions at potential synonymous sites (dS) and the dN/dS ratios were assessed to estimate the selective pressure during the follow-up period on circulating HIV-1 RNA from each randomly selected children or adolescents. No difference in mean dN could be observed between (I+, VF+) and (I-, VF+) subgroups for all pairwise comparisons of HIV-1 pol sequences between inclusion and follow-up (15.9 ± 11.7 in (I+, VF+) subgroup vs. 13.2 ± 7.2 in (I-, VF+) subgroup; P > 0.05). In contrast, mean dS was 1.8-fold higher in (I+, VF+) than (I-, VF+) subgroups (25.9 ± 18.4 in (I+, VF+) subgroup vs. 14.3 ± 10.8 in (I-, VF+) subgroup; P < 0.05). In the (I+, VF+) subgroup, the mean dS was 1.6-fold higher than the mean dN (P < 0.01). Finally, the mean ω ratio was 2.1-fold lower in (I+, VF+) than (I-, VF+) subgroups (0.6 ± 0.3 in (I+, VF+) subgroup vs. 1.3 ± 0.7 in (I-, VF+) subgroup; P < 0.001), indicating purifying selection in the immunovirological discordant (I+, VF+) subgroup and positive selection in the immunovirological failure (I-, VF+) subgroup.
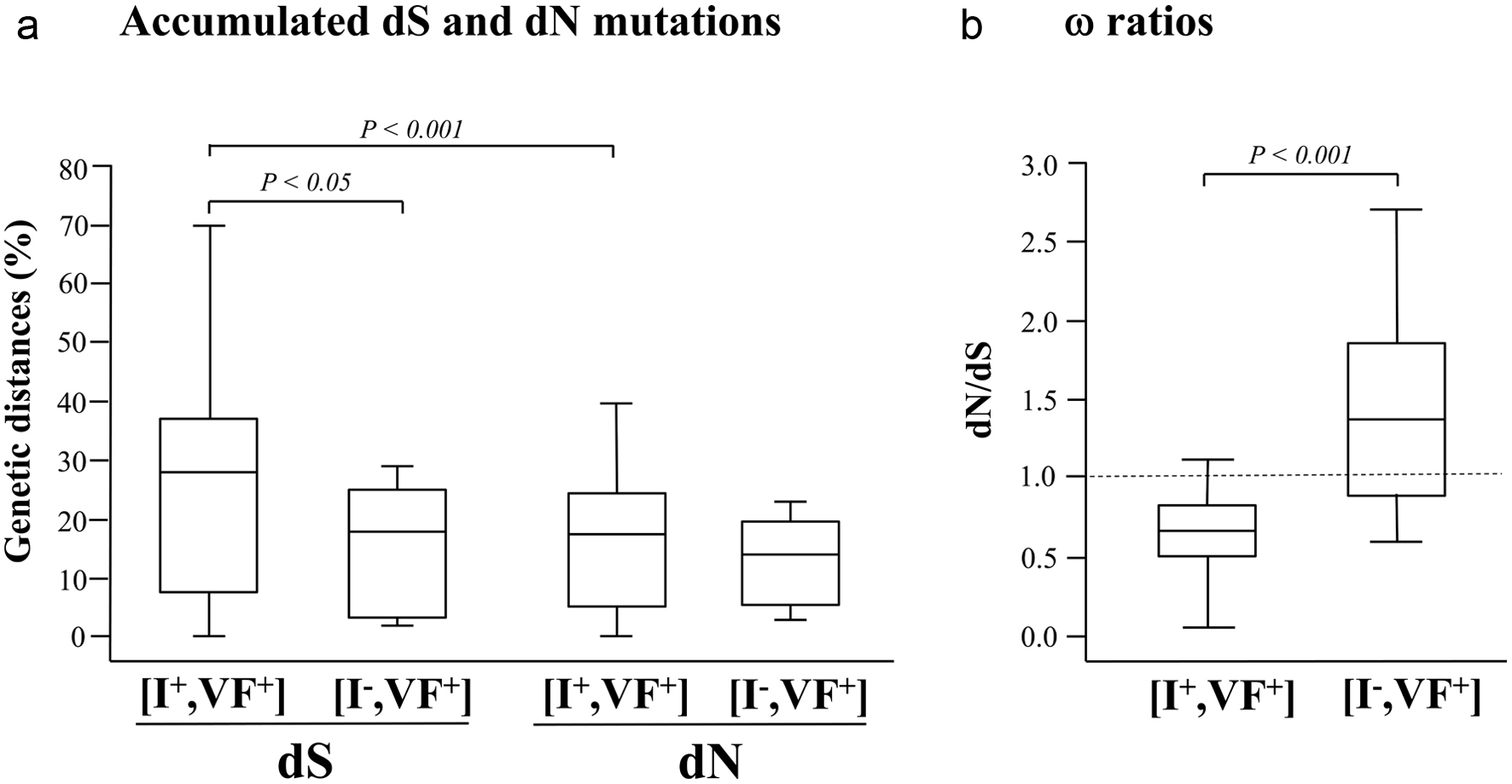 Click for large image | Figure 2. Estimates of codon-based evolutionary divergences between pairs of HIV-1 pol gene nucleotide sequences during a 39 months period of prospective follow-up of children and adolescents attending the Complexe Pediatrique of Bangui, Central African Republic. The genetic distances (expressed in percentage) of synonymous substitutions per 100 potential synonymous sites (dS) and of nonsynonymous substitutions per 100 potential nonsynonymous site (dN) accumulated during the follow-up period in circulating HIV-1 pol gene (a), as well as the (dN/dS) ω ratios (b) are shown for both (I+, VF+) and (I-, VF+) subgroups. For each distribution, the blank box represents the interquartile ranges, the horizontal bar corresponds to the median and the vertical bars and hats indicate the 10th and 90th percentiles. The dotted line identifies the neutral selection ratio (ω = 1). Comparisons used Wilcoxon’s test for paired data and Welch’s t-test between subgroups. P values are given above indentations. |
| Discussion | ▴Top |
Immunological surrogate marker (CD4 T cell count) and HIV-1 genetic diversity in pol gene were evaluated longitudinally in perinatally HIV-1-infected children and adolescents born from HIV-infected mothers, in virological failure despite long-term WHO-recommended ART and followed up for 39 months at the Complexe Pediatrique of Bangui. The immunological responses to ART corresponded basically to two categories, including full therapeutic failure with immunological and virological nonresponses in nearly one-quarter of children always associated with high levels of DRMs, and discordant immunovirological responses in the majority (about 75%) of remaining children. Outstandingly, the high prevalence of children and adolescents harboring discordant immunovirological responses with paradoxical high CD4 T cell counts and high HIV-1 RNA load demonstrates a posteriori minimal correlation between immunological and virological biological markers.
The subgroup of children and adolescents in full therapeutic failure (I-, VF+) in whom plasma HIV-1 RNA load was elevated overtime and CD4 T cell counts remained persistently low after the follow-up period represented one-quarter of children and adolescents in virological failure, in keeping with generally high rate of ART failure in African pediatric cohorts [2, 21-26].
The subgroup of children and adolescents who exhibited discordant response to treatment with immunological response and virological nonresponse (I+, VF+) represented the majority of cohort children in virological failure. Although these patients failed to control HIV-1 viral replication after the follow-up period and thus were all in virological failure with viruses harboring high rates of DRMs (> 97%), their CD4 T cell counts have remained persistently above normal levels according to age of children (i.e., CD4 T cells count > 750/µL for children less than 5 years and CD4 T cells count > 500/µL for children and adolescents older than 5 years, as defined by the WHO) [3, 4]. Increased CD4 T cell count, higher than the WHO-threshold of immunological success without suppressing viral load demonstrating discordant response, was previously reported [7, 26-30]. It has been suggested that different mechanisms may be involved in this phenomenon. Firstly, the recovery of thymic function and higher thymic output under ART favor immune restoration in HIV-1-infected children [27, 28, 31-33]. Secondly, partial virus suppression under certain drugs regimen (mainly including PI) may reduce CD4 T cell turnover and activation [24, 34]. Thirdly, the resistance mutation V82A (in protease) previously reported in discordant patients in virological failure but with persistent high levels of CD4 counts [20], was observed in a minority of children in the (I+, VF+) subgroup. Fourthly, discordant responses during ART may be related to enhance HIV-directed immune responses, diminished cellular activation, and preservation of non-syncytium-inducing (CCR5-tropic) viruses, as previously reported [30]. Fifthly, the accumulation of high level of DRMs overtime could have provided impaired viral fitness [28, 35-40], which would substantially modify the viral pathogenic potential [41]. Finally, persistent normal or elevated CD4 T cell count associated with high circulating HIV-1 RNA load may procure selective advantages for the virus as well as for the discordant children, who appeared unexpectedly tolerant to the virus.
The evolutionary patterns of HIV-1 pol gene diversity were further evaluated in the two categories of children and adolescent in virological failure by pairwise comparisons of the numbers of synonymous (silent) and nonsynonymous (amino acid altering) nucleotide substitutions in HIV-1 pol gene sequences between inclusion and follow-up. Amino acid sites in viral proteins are expected to be under different selective pressures. Thus, the genetic distances of synonymous amino acids (e.g., dS) reflect mutation rates and generation times, whereas the distances of nonsynonymous (e.g., dN) may be affected by selective factors, especially immune pressure [42], reflecting the adaptive evolution against the background rate of evolution [43]. Remarkably, the evolutionary patterns of HIV-1 pol gene diversity in the study cohort differed between the two categories of virological failure, with different underlying ω (dN/dS) ratios, pointing to the existence of different viruses and host factors contributing to the evolution of HIV-1 quasispecies in vivo. These findings likely denote the lack of genetic homogeneity between circulating HIV-1 RNA from the two groups of children in virological failure, with concordant (I-, VF+) or discordant (I+, VF+) immunological response. Different evolutionary patterns mainly in HIV-1 env gene were previously reported during the natural history of HIV-1 infection as well as during ART [29, 42, 44-46].
Within children and adolescents in full therapeutic failure, the mean ω ratio of viral strains between baseline and follow-up was above 1, suggesting diversifying positive selection in HIV-1 pol gene [19]. This observation contrasts with natural history of HIV-1 infection in the absence of ART during which the rate of synonymous substitution in HIV-1 pol gene generally exceeds that of nonsynonymous substitution, indicating that pol gene is naturally subject to purifying selection [44]. Positive selection of HIV-1 pol-encoded amino acids overtime in patients in virological failure under ART may affect viral proteins conformations and/or biological functions, possibly to escape to antiretroviral drug pressure. Despite the low CD4 T cell counts in the (I-, VF+) subgroup, another hypothesis is that positive selection in pol gene may occur under the selective pressure of changes in the dynamics of the HIV-specific immune response. Indeed, positive selection in HIV-1 env gene with increase in dN/dS ratio in the V3 loop and increase of dN/dS ratio above 1 in genes coding for cytotoxic T lymphocytes epitopes in HIV-1 variants has been hypothesized to be caused by immune-mediated selection for sequence diversity in order to avoid immune recognition [44, 47, 48].
Within children and adolescents with persistent discordant immunological response during virological failure under prolonged ART, the mean ω ratio of viral strains between baseline and follow-up was below 1, suggesting purifying selection overtime in HIV-1 pol gene sequences in the (I+, VF+) subgroup [19, 42]. In addition, viral strains in (I+, VF+) subgroup accumulated synonymous mutations overtime. These findings indicate neutral or relatively limited genetic evolution and likely suggest genetic adaptation of the viruses to host functional constraints, similarly to the natural selection on HIV-1 pol gene in the absence of ART, which is basically subject to purifying selection [44], suggesting that genetic drift rather than deterministic pressure is likely to play a substantial role in viral evolution in immunovirological discordant children and adolescents. Furthermore, our observations about the accumulation of synonymous substitutions in circulating HIV-1 RNA pol gene sequences with their purifying selection overtime in the (I+, VF+) subgroup also support the hypothesis that persistent replication of HIV-1 strains with high level of DRMs and probably modified viral fitness could be largely independent of HIV-specific immune response. Finally, faster progression to AIDS was previously correlated with higher rate of synonymous substitutions [46], and similar hypothesis in the pediatric cohort at the Complexe Pediatrique of Bangui warrants further investigation.
The discordant immunovirological responses in our study in perinatally HIV-infected children are reminiscent of the pediatric non-progressor children described by the group of Phillip Goulder in South Africa [49]. These children expressed two cardinal immunological features of non-pathogenic simian immunodeficiency virus (SIV) infection in sooty mangabeys, low immune activation despite high viremia and low CCR5 expression on long-lived central memory CD4 T-cells [49].
In conclusion, our observations provide important information on immunological outcome of perinatally infected children and adolescents in virological failure despite receiving lifelong WHO-recommended ART. Thus, very particular host-pathogen interactions occur in HIV-infected children and adolescents born from HIV-infected mother harboring sustained HIV replication under long-term ART. In the (I-, VF+) subgroup of children and adolescents in full therapeutic failure, the positive selection is strong, suggesting negligible genetic drift and deterministic evolution under therapeutic and host factors. In contrast, in the (I+, VF+) subgroup of children and adolescents of discordant immunological response and virological failure, the viral evolution would likely proceed according to a stochastic model of genetic drift.
Acknowledgments
We are particularly grateful to Dr. Alexis Naissem and Mr. Dionke Fofana, from Ensemble pour une Solidarite Therapeutique en Reseau (GIP-ESTHER, Paris, France) and for the Expertise France, Paris, for contributions and relevant discussions. We thank Miss Rosine Feissona for excellent technical assistance.
Financial Disclosure
The authors declare that they had received no funding regarded the performing of this study.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Informed Consent
Informed written consent was obtained from mothers for themselves and on behalf of their respective child.
Author Contributions
RSMB, CDMK, JCG, and LB conceived and designed the experiments. CDMK, LB, and RSMB analyzed the data. PR and RSMB carried out resistance genotypes and phylogenetic analyses. GG and JCG contributed to patients’ data collection and ethical committee approval. CC, CDMK, DV, HP, JFDS, LB, MAJ, PR, RSMB and SM wrote the paper. All authors read and approved the final version of the manuscript.
Data Availability
The data supporting the findings of this study have been deposited in the European Nucleotide Archive at GenBankEMBL-EBI (available at: http://www.ebi.ac.uk/ena/data/view/) and can be accessed with the following accession numbers: LT577626-LT577673; LT578170-LT578280 and LT726745-LT726792.
| References | ▴Top |
- UNAIDS. Global HIV & AIDS statistics - 2018 fact sheet. Available at: http://www.unaids.org/en/resources/fact-sheet (Last accessed March 27, 2020).
- Jenabian MA, Costiniuk CT, Mboumba Bouassa RS, Chapdeleine Mekue Mouafo L, Brogan TV, Belec L. Tackling virological failure in HIV-infected children living in Africa. Expert Rev Anti Infect Ther. 2015;13(10):1213-1223.
doi pubmed - World Health Organization (WHO) recommendations. Antiretroviral therapy for HIV infection in adults and adolescents. Antiretroviral therapy for a public health approach 2010 revision. Available at: http://whqlibdoc.who.int/publications/2010/9789241599764_eng.pdf (Last accessed June 1, 2016).
- World Health Organization (WHO, 2013) recommendations. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. June 2013. Available at: http://www.who.int/hiv/pub/guidelines/arv2013/download/en/ (Last accessed April 01, 2020).
- World Health Organization (WHO, 2017). HIV treatment and care. What's new in treatment monitoring: viral load and CD4 testing. Update July 2017. Available at: http://apps.who.int/iris/bitstream/handle/10665/255891/WHO-HIV-2017.22-eng.pdf;jsessionid=22AA151AECD9CD87D39B57B98F710CE1?sequence=1 (Last accessed April 01, 2020).
- World Health Organization (WHO, 2014) supplement. March 2014 supplement to the 2013 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach. Available at: https://www.who.int/hiv/pub/guidelines/arv2013/arvs2013upplement_march2014/en/ (Last accessed April 01, 2020).
- World Health Organization (WHO, 2016). Guidelines. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach. Second Edition 2016. p. 272. Available at: http://apps.who.int/iris/bitstream/10665/208825/1/9789241549684_eng.pdf (Last accessed April 01, 2020).
- World Health Organization (WHO, 2018). Updated recommendations on first-line and second-line antiretroviral regimens and post-exposure prophylaxis and recommendations on early infant diagnosis of HIV: interim guidelines. Available at: https://apps.who.int/iris/bitstream/handle/10665/277395/WHO-CDS-HIV-18.51-eng.pdf?ua=1 (Last accessed April 01, 2020).
- Lawn SD, Little F, Bekker LG, Kaplan R, Campbel E, Orrell C, Wood R. Changing mortality risk associated with CD4 cell response to antiretroviral therapy in South Africa. AIDS. 2009;23(3):335-342.
doi pubmed - Barth RE, Tempelman HA, Smelt E, Wensing AM, Hoepelman AI, Geelen SP. Long-term outcome of children receiving antiretroviral treatment in rural South Africa: substantial virologic failure on first-line treatment. Pediatr Infect Dis J. 2011;30(1):52-56.
doi pubmed - Schoffelen AF, Wensing AM, Tempelman HA, Geelen SP, Hoepelman AI, Barth RE. Sustained virological response on second-line antiretroviral therapy following virological failure in HIV-infected patients in rural South Africa. PLoS One. 2013;8(3):e58526.
doi pubmed - Gody JC, Charpentier C, Mbitikon O, Si-Mohamed A, LeGoff J, Gresenguet G, Belec L. High prevalence of antiretroviral drug resistance mutations in HIV-1 non-B subtype strains from African children receiving antiretroviral therapy regimen according to the 2006 revised WHO recommendations. J Acquir Immune Defic Syndr. 2008;49(5):566-569.
doi pubmed - Charpentier C, Gody JC, Mbitikon O, Moussa S, Matta M, Pere H, Fournier J, et al. Virological response and resistance profiles after 18 to 30 months of first- or second-/third-line antiretroviral treatment: a cross-sectional evaluation in HIV type 1-infected children living in the Central African Republic. AIDS Res Hum Retroviruses. 2012;28(1):87-94.
doi pubmed - Mossoro-Kpinde CD, Gody JC, Mboumba Bouassa RS, Mbitikon O, Jenabian MA, Robin L, Matta M, et al. High levels of virological failure with major genotypic resistance mutations in HIV-1-infected children after 5 years of care according to WHO-recommended 1st-line and 2nd-line antiretroviral regimens in the Central African Republic: A cross-sectional study. Medicine (Baltimore). 2017;96(10):e6282.
doi pubmed - Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418-426.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731-2739.
doi pubmed - Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267(5608):275-276.
doi pubmed - Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111-120.
doi pubmed - Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155(1):431-449.
- Natesampillai S, Nie Z, Cummins NW, Jochmans D, Bren GD, Angel JB, Badley AD. Patients with discordant responses to antiretroviral therapy have impaired killing of HIV-infected T cells. PLoS Pathog. 2010;6(11):e1001213.
doi pubmed - Germanaud D, Derache A, Traore M, Madec Y, Toure S, Dicko F, Coulibaly H, et al. Level of viral load and antiretroviral resistance after 6 months of non-nucleoside reverse transcriptase inhibitor first-line treatment in HIV-1-infected children in Mali. J Antimicrob Chemother. 2010;65(1):118-124.
doi pubmed - Kebe K, Thiam M, Diagne Gueye NR, Diop H, Dia A, Signate Sy H, Charpentier C, et al. High rate of antiretroviral drug resistance mutations in HIV type 1-infected Senegalese children in virological failure on first-line treatment according to the World Health Organization guidelines. AIDS Res Hum Retroviruses. 2013;29(2):242-249.
doi pubmed - Salou M, Dagnra AY, Butel C, Vidal N, Serrano L, Takassi E, Konou AA, et al. High rates of virological failure and drug resistance in perinatally HIV-1-infected children and adolescents receiving lifelong antiretroviral therapy in routine clinics in Togo. J Int AIDS Soc. 2016;19(1):20683.
doi pubmed - Deeks SG, Hoh R, Grant RM, Wrin T, Barbour JD, Narvaez A, Cesar D, et al. CD4+ T cell kinetics and activation in human immunodeficiency virus-infected patients who remain viremic despite long-term treatment with protease inhibitor-based therapy. J Infect Dis. 2002;185(3):315-323.
doi pubmed - Bandera A, Ferrario G, Saresella M, Marventano I, Soria A, Zanini F, Sabbatini F, et al. CD4+ T cell depletion, immune activation and increased production of regulatory T cells in the thymus of HIV-infected individuals. PLoS One. 2010;5(5):e10788.
doi pubmed - Kelly C, Gaskell KM, Richardson M, Klein N, Garner P, MacPherson P. Discordant immune response with antiretroviral therapy in HIV-1: a systematic review of clinical outcomes. PLoS One. 2016;11(6):e0156099.
doi pubmed - Chavan S, Bennuri B, Kharbanda M, Chandrasekaran A, Bakshi S, Pahwa S. Evaluation of T cell receptor gene rearrangement excision circles after antiretroviral therapy in children infected with human immunodeficiency virus. J Infect Dis. 2001;183(10):1445-1454.
doi pubmed - Ometto L, De Forni D, Patiri F, Trouplin V, Mammano F, Giacomet V, Giaquinto C, et al. Immune reconstitution in HIV-1-infected children on antiretroviral therapy: role of thymic output and viral fitness. AIDS. 2002;16(6):839-849.
doi pubmed - Bagnarelli P, Vecchi M, Burighel N, Bellanova D, Menzo S, Clementi M, De Rossi A. Genotypic and phenotypic correlates of the HIV Type 1 env gene evolution in infected children with discordant response to antiretroviral therapy. AIDS Res Hum Retroviruses. 2004;20(12):1306-1313.
doi pubmed - Sufka SA, Ferrari G, Gryszowka VE, Wrin T, Fiscus SA, Tomaras GD, Staats HF, et al. Prolonged CD4+ cell/virus load discordance during treatment with protease inhibitor-based highly active antiretroviral therapy: immune response and viral control. J Infect Dis. 2003;187(7):1027-1037.
doi pubmed - Douek DC, Koup RA, McFarland RD, Sullivan JL, Luzuriaga K. Effect of HIV on thymic function before and after antiretroviral therapy in children. J Infect Dis. 2000;181(4):1479-1482.
doi pubmed - De Rossi A, Walker AS, Klein N, De Forni D, King D, Gibb DM. Increased thymic output after initiation of antiretroviral therapy in human immunodeficiency virus type 1-infected children in the Paediatric European Network for Treatment of AIDS (PENTA) 5 Trial. J Infect Dis. 2002;186(3):312-320.
doi pubmed - De Rossi A. Virological and immunological response to antiretroviral therapy in HIV-1 infected children: genotypic and phenotypic assays in monitoring virological failure. New Microbiol. 2004;27(2 Suppl 1):45-50.
- Deeks SG, Barbour JD, Grant RM, Martin JN. Duration and predictors of CD4 T-cell gains in patients who continue combination therapy despite detectable plasma viremia. AIDS. 2002;16(2):201-207.
doi pubmed - Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72(9):7632-7637.
doi pubmed - Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72(4):3300-3306.
doi pubmed - Mammano F, Trouplin V, Zennou V, Clavel F. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J Virol. 2000;74(18):8524-8531.
doi pubmed - Menzo S, Monachetti A, Balotta C, Corvasce S, Rusconi S, Paolucci S, Baldanti F, et al. Processivity and drug-dependence of HIV-1 protease: determinants of viral fitness in variants resistant to protease inhibitors. AIDS. 2003;17(5):663-671.
doi pubmed - Prado JG, Parkin NT, Clotet B, Ruiz L, Martinez-Picado J. HIV type 1 fitness evolution in antiretroviral-experienced patients with sustained CD4+ T cell counts but persistent virologic failure. Clin Infect Dis. 2005;41(5):729-737.
doi pubmed - Martinez-Picado J, Martinez MA. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. 2008;134(1-2):104-123.
doi pubmed - Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151-178.
doi pubmed - Edwards CT, Holmes EC, Pybus OG, Wilson DJ, Viscidi RP, Abrams EJ, Phillips RE, et al. Evolution of the human immunodeficiency virus envelope gene is dominated by purifying selection. Genetics. 2006;174(3):1441-1453.
doi pubmed - Miyata T, Yasunaga T. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J Mol Evol. 1980;16(1):23-36.
doi pubmed - Seibert SA, Howell CY, Hughes MK, Hughes AL. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol Biol Evol. 1995;12(5):803-813.
- Casado C, Garcia S, Rodriguez C, Del Romero J, Bello G, Lopez-Galindez C. Different evolutionary patterns are found within human immunodeficiency virus type 1-infected patients. J Gen Virol. 2001;82(Pt 10):2495-2508.
doi pubmed - Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, et al. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3(2):e29.
doi pubmed - Bonhoeffer S, Holmes EC, Nowak MA. Causes of HIV diversity. Nature. 1995;376(6536):125.
doi pubmed - Nielsen R. Changes in ds/dn in the HIV-1 env gene. Mol Biol Evol. 1999;16(5):711-714.
doi pubmed - Muenchhoff M, Adland E, Karimanzira O, Crowther C, Pace M, Csala A, Leitman E, et al. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Sci Transl Med. 2016;8(358):358ra125.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Clinical Medicine Research is published by Elmer Press Inc.


