| Journal of Clinical Medicine Research, ISSN 1918-3003 print, 1918-3011 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Clin Med Res and Elmer Press Inc |
| Journal website https://www.jocmr.org |
Original Article
Volume 14, Number 10, October 2022, pages 416-424
STAT-5 and STAT-6 in Breast Cancer: Potential Crosstalk With Estrogen and Progesterone Receptors Can Affect Cell Proliferation and Metastasis
Mohamed Oueslatia, d, Ilhem Bettaiebb, Ridha Ben Younesa, Amor Gamoudib, Khaled Rahalc, Ridha Oueslatia
aUnit of Immunology, Environmental Microbiology and Carcinogenesis (IMEC), Faculty of Sciences of Bizerte, University of Carthage, Zarzouna 7021, Tunisia
bDepartment of Immuno-Histo-Cytology, Salah Azaiz Cancer Institute, Tunis, Tunisia
cDepartment of Carcinologic Surgery, Salah Azaiz Cancer Institute, Tunis, Tunisia
dCorresponding Author: Mohamed Oueslati, Unit of Immunology, Environmental Microbiology and Carcinogenesis (IMEC), Faculty of Sciences of Bizerte, University of Carthage, Zarzouna 7021, Tunisia
Manuscript submitted July 13, 2022, accepted September 5, 2022, published online October 28, 2022
Short title: STAT5a and STAT6 Effects in Breast Cancer
doi: https://doi.org/10.14740/jocmr4785
| Abstract | ▴Top |
Background: Signal transducers and activators of transcription 5a and 6 (STAT5a and STAT6) play a critical role in tumorigenesis of mammary glands. Based on previous studies, the breast cancer is largely dependent on hormone receptors. Consequently, it is very interesting to decipher the relationship between the STAT5a and STAT6 expression and the molecular distribution of estrogen receptors (ERs) and progesterone receptors (PRs) in mammary tumors.
Methods: Our study analyzed the expression of STAT5a and STAT6, ERα, ERβ and PR in 40 breast tumor tissues using quantitative real-time polymerase chain reaction (qRT-PCR). Furthermore, the Ki-67 and HER2 status were detected using immunohistochemistry.
Results: STAT5a and STAT6 were retained in the majority of the cases studied. Increasing of STAT5a and STAT6 is significantly associated with ERs and PR. The coexpression of both STAT5a and STAT6 with ERs and PR is associated with high tumor grades. Moreover, the coexpression of STAT5a and STAT6 with ERα and PR is associated with a high proliferation index. In addition, (STAT6 + ERβ+) and (STAT6 + PR+) breast cancer subgroups are associated with lymph node infiltration (P = 0.001 and P = 0.03, respectively).
Conclusions: Our study results provide an interaction between STAT5a and STAT6 with ERs and PR inducing cell proliferation. Coexpression of STAT5a and STAT6 with ERs and PR can predict sensibility to hormonal therapy.
Keywords: Breast cancer; STAT5a; STAT6; ERs; PR; Crosstalk
| Introduction | ▴Top |
Mammary gland development occurs through the activation of a variety of transcription factors. Inappropriate or constitutive activation of many of these transcription factors is found in breast cancer and may contribute directly to its pathogenesis [1]. Particularly, signal transducers and activators of transcription (STAT), a family of transcription factors, play crucial roles in many cellular functions and are often activated unsuitably in cancer [2]. STATs are latent transcription factors that localized in the cytoplasm. Upon their activation by tyrosine phosphorylation, STATs can dimerize, translocate to the nucleus, bind to DNA, and modulate transcription. Thereby, they regulate cellular functions such as survival, proliferation, and differentiation [3]. Two STAT family members, STAT5 and STAT6, play important roles in normal mammary gland development and both have been implicated in breast tumorigenesis [4].
STAT5, which includes two homologous proteins, STAT5a and STAT5b, plays a crucial role in normal mammary gland development and carcinogenesis. Indeed, STAT5 is activated during post-pubertal mammary gland development, but it was observed to cause epithelial hyper-proliferation and generates precocious functional alveoli formation in virgin mice [5]. Deficient STAT5 mice exhibit a lack of lobulo-alveolar growth of mammary glands and females are unable to lactate [6]. In addition, STAT-5a mediates the prolactin-induced milk protein gene transcription during lactation [7]. The immunohistochemistry study in a large cohort of malignant breast tumors shows that STAT5 was constitutively activated [8]. In addition, Cotarla et al (2004) found that STAT5a is activated in a high proportion of breast cancers [9]. In fact, STAT5a is phosphorylated in nearly 70% of human malignant breast tumors. Furthermore, murine models also support the idea of the role played by STAT5 in mammary tumorigenesis. Mice that express a constitutively activated form of STAT5 develop mammary carcinomas, whereas mice that lack STAT5a are protected against mammary tumors induced by transforming the growth factor α [10]. Taken together, these data imply that STAT5 plays a main role in both normal and neoplastic mammary functions.
STAT6 also plays a critical role in mammary gland development and in breast cancer. During early pregnancy, in response to the upregulation of Th2 cytokines (interleukin (IL)-4 and IL-13), STAT6 stimulates alveolar differentiation and proliferation through the induction of the GATA3 expression [11]. STAT6 is instrumental in regulating the balance between Th1 and Th2 cells. It promotes tumor invasiveness and metastasis through the promotion of the Th2 cytokines profile. Moreover, STAT6 can inhibit immunosurveillance against primary solid tumors and metastatic disease. In fact, STAT6-deficient mice are resistant to mammary carcinoma and reject metastatic immortal tumor cells [12].
On the other hand, normal mammary development and breast cancer are largely dependent on other factors such as estrogen receptors (ERs) and progesterone receptors (PRs). In fact, ERs and PRs are critical mediators for mammary gland proliferation after puberty [13]. Furthermore, ERs and PRs may induce cell proliferation and metastasis in breast cancer [14-16]. The present study, therefore, attempts to focus on the assessment of the STAT5a and STAT6 expression levels in malignant breast tumors according to ER and PR distribution.
| Materials and Methods | ▴Top |
Patients and samples
Malignant mammary tumors were excised during tumorectomies from 40 women (mean age 58.5, range 32 - 85 years) at the Saleh Azaiez Oncology Institute of Tunisia. The women did not undergo any treatment before surgery. The presence of malignant cells in the samples was determined by a pathology specialist. Each diagnosed sample was divided into two portions: one portion was immediately processed for immunohistochemistry and the other portion was frozen and maintained at - 80 °C until RNA extraction. All the procedures followed were examined and approved by the Saleh Azaiez Oncology Institute Ethics Committee. The study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
RNA extraction
RNA was extracted from breast specimens by using a mechanical stirrer in the presence of a lysis buffer prior to the use of the total RNA isolation and high pure RNA isolation kits (Roche Diagnostics, Basel, Switzerland) according to the manufacturer’s protocol.
cDNA synthesis
The mRNA concentration was measured with a NanoDrop spectrophotometer, the cDNA synthesis was done in a total volume of 20 µL using the PrimeScript™ first strand cDNA Synthesis Kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacture’s protocol. Mix1 was prepared containing 1 µg of RNA, 2 µL of random hexamer primers, 1 µL of dNTP mixture and RNAse-free water (total volume 10 µL). The samples were then incubated for 5 min at 65 °C. Subsequently, Mix2 was prepared containing Mix1, 4 µL of 5 × PrimScript buffer, 0.5 µL of RNase inhibitor, 1 µL of PrimScriptRTase and 4.5 µL of RNAse-free water (total volume 20 µL). The samples were incubated at 30 °C for 10 min and at 42 °C for 60 min, and reverse transcriptase were inactivated by heating at 95 °C for 5 min.
PCR amplification
Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out using an ABI Prism 7700 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Specific primers were STAT5a-forward, 5’-CACAGATCAAGCAAGTGGTC-3’; STAT5a-reverse, 5’-CTGTCCATTGGTCGGCGTAA-3’ and STAT6-forward, 5’-CCTCGTCACCAGTTGCTT-3’; STAT6-reverse, 5’-TCCAGTGCTTTCTGCTCC-3’. The 36B4 expression was used as a control to normalize the data. Primer sequences used to amplify 36B4 were 5’-AATCTCCAGGGGCACCATT-3’ and reverse 5’-CGCTGGCTCCCACTTTGT-3’. Relative mRNA levels were calculated based on the CT values and corrected for the 36b4 expression according to the equation 2-ΔΔCT [17]. Relative mRNA levels in the control tissue were equated to 1 and the other values were expressed relative to this. Experiments were performed in triplicate for each data point.
Immunohistochemistry
The immunohistochemistry expression of the oncoprotein Her2/neu and the proliferation index Ki67 were tested on the same set of tumors. The primary antibodies used were: mouse anti-human Her2 (CB11) and mouse anti-human Ki67 (MM1) (NovoCastra, Newcastle, UK). After deparaffinization Sections were subsequently hydrated, incubated for 30 min in 1% hydrogen peroxide to block endogenous activity, and then antigen retrieval was performed by incubating the sections in a 0.01 M citrate buffer (epitope retrieval solution pH 6.0; Leica Microsystems GmbH, Wetzlar, Germany) for 30 min at 98 °C. The primary antibodies were applied for 1 h at 4 °C, with a dilution of 1:40 for Her2 and 1:200 for Ki-67. The sections were then incubated at room temperature with post primary block for 30 min to block nonspecific polymer binding. The sections were incubated with a NovoLink™ Polymer for 30 min at room temperature, followed by incubations with 3,3’-diaminobenzidine (DAB) working solution for 5 min at room temperature to develop peroxidase activity. The slides were counterstained with hematoxylin and mounted. Staining specificity was checked using negative controls. Subsequently, the primary antibodies were applied for 1 h at 4 °C, with a dilution of 1:40 for Her2 and 1:200 for Ki-67. The sections were then incubated at room temperature with post primary block for 30 min to block nonspecific polymer binding. The sections were incubated with a NovoLink™ Polymer for 30 min at room temperature, followed by incubations with 3,3’-diaminobenzidine (DAB) working solution for 5 min at room temperature to develop peroxidase activity. The slides were counterstained with hematoxylin and mounted. Staining specificity was checked using negative controls. This part has been previously described by Oueslati et al, 2017 [18].
Statistical analysis
The Chi-square test with the R (i386 3.2.1) software was used to compare the quantitative PCR results of STAT5a and STAT6 with clinic-pathological characteristics and with the ERs and PR status. The t-test and Mann-Whitney test with GraphPad Prism software were used to compare the STAT5a and STAT6 mRNA level with the clinical and molecular settings. Data are presented as the mean ± standard deviation. P < 0.05 was considered to indicate a statistically significant difference.
| Results | ▴Top |
STAT5a and STAT6 mRNA expressions and their relationship with clinical and molecular settings
STAT5 and STAT6 have both been reported to be activated in breast tumors and also to have distinct roles in the normal mammary gland and in cancerogenesis. On the other hand, the normal and the pathological mammary glands strongly depend on hormonal receptors. Consequently, in this paper the STAT5a and STAT6 mRNA expressions were evaluated using the qRT-PCR technique in a cohort of malign breast tumors as well as their relationship with ERs and PRs. Our results show a high positivity of both STAT5a and STAT6 (Table 1). The STAT5a and STAT6 expressions were not significantly linked to the menopausal status. The analysis of the mRNA expression level of STAT5a and STAT6 revealed a significant increase of STAT5a in postmenopausal breast tumors (P = 0.03) (Fig. 1). On the contrary, the STAT6 mRNA expression level was three times higher in premenopausal breast tumors compared to postmenopausal breast tumors (P = 0.02) (Fig. 1). When compared to the tumor grade, there was a significant association between the STAT5a expression and the Scarff-Bloom-Richardson (SBR)II tumor grade (P = 0.0005) (Table 1); while STAT6 is associated with a high tumor grade (SBRII and SBRIII) (P = 0.002) (Table 1).
 Click to view | Table 1. Relationship Between the mRNA STAT5a and STAT6 Expressions and the Standard Clinical and Pathological Factors and Molecular Settings |
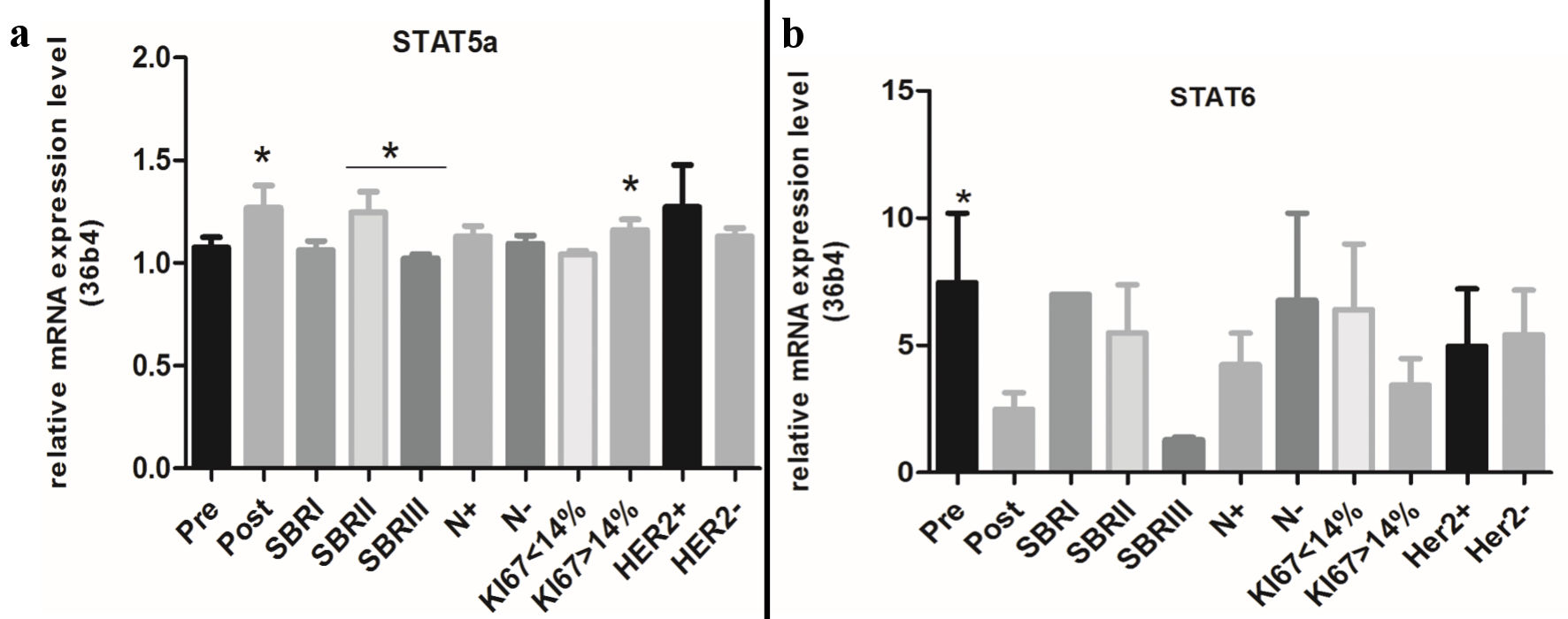 Click for large image | Figure 1. Relative mRNA expression levels of STAT5a and STAT6 according to clinical and molecular information. STAT: signal transducers and activators of transcription; SBR: Scarff-Bloom-Richardson. |
The analysis of the STAT5a mRNA level shows a significant difference between the SBRII and SBRIII tumor grades (P = 0.01) (Fig. 1). Our results summarized in Table 2 also show a significant association of the STAT6 (not STAT5a) expression with node infiltration (metastasis) (P = 0.01).
 Click to view | Table 2. Relationship Between the STAT5a, STAT6 and ERα, ERβ and PR Expressions in Mammary Malign Tumors |
Both STAT5a and STAT6 were correlated with a tumor size smaller than 30 mm (P = 0.0005 and P = 0.0001, respectively). A significant negative association of the STAT5a and STAT6 overexpressions with the HER2/neu status (P = 0.003 and P = 0.004, respectively) was also found but not with the mRNA levels of STAT5a and STAT6. For Ki-67 (Fig. 2) there is not a significant association of the STAT5a and STAT6 expressions with the proliferation index; however, our results show an association between the increase of the STAT5a mRNA level (but not for STAT6) with a Ki-67 index > 14% (P = 0.04) (Fig. 1).
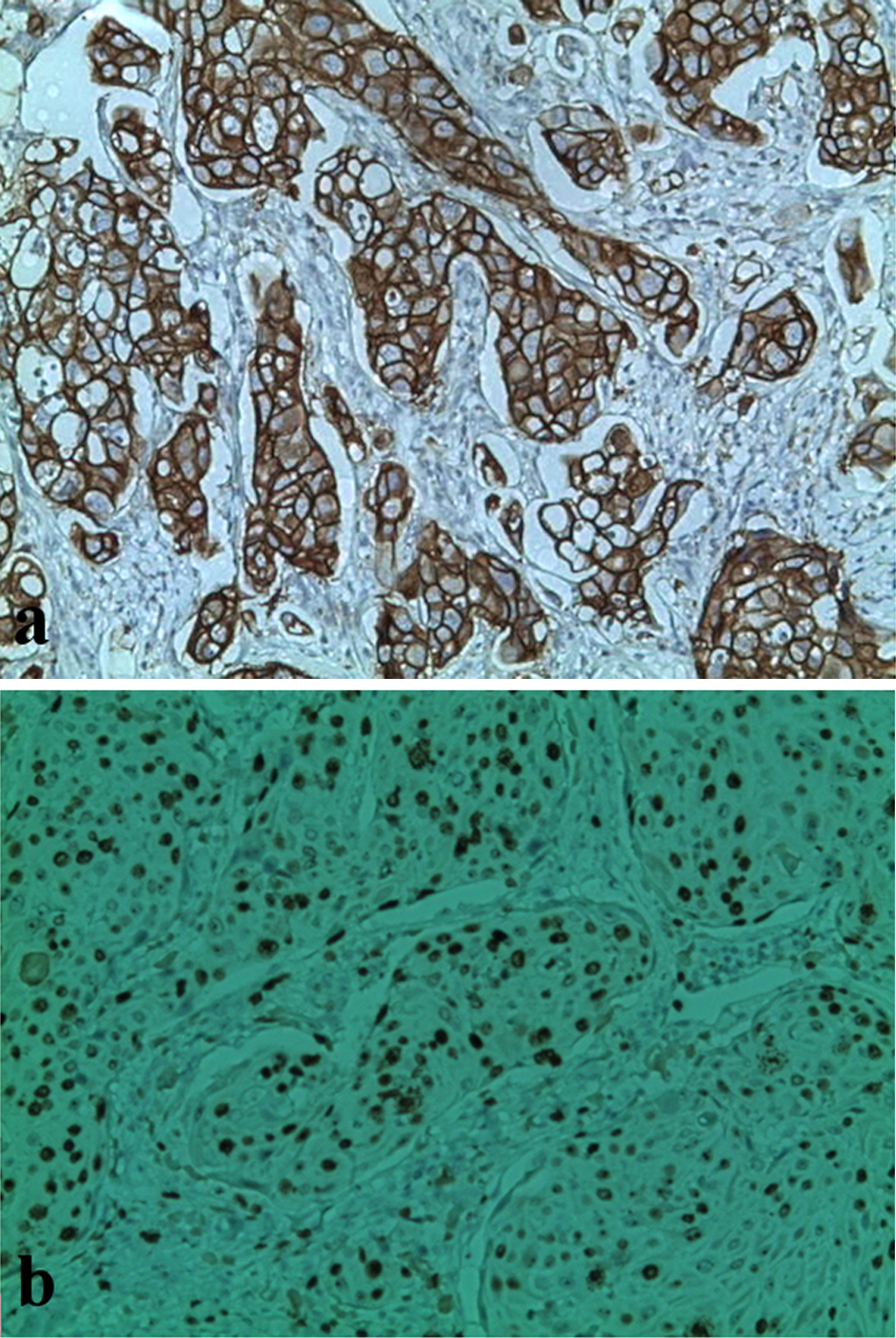 Click for large image | Figure 2. Immunochemical staining with anti-HER2 and anti-Ki-67 antibodies in breast tumors. (a) Presence of overexpression of HER2 oncoprotein (HER2 score: 3; original magnification, × 400). (b) Ki-67 proliferation index estimated at 70% (original magnification, × 250). |
Relationship between the STAT5a and STAT6 expressions and the ERα, ERβ and PR status
In order to determine the relationship between the STAT5a and STAT6 expressions and hormonal receptors, the coexpression was analyzed (Table 2). The STAT5a and STAT6 expressions are significantly associated (P = 0.01 and P = 0.02, respectively) with ERα+ (but not with ERβ+). Furthermore, the STAT5a and STAT6 expressions are strongly associated with PR+ (P = 0.000061 and P = 0.00000606, respectively) (Table 2).
Relationship between (STAT5a + ERα+), (STAT5a + ERβ+) and (STAT5a + PR+) breast cancer subgroups and clinical settings
Our results (Table 3) reveal the absence of a significant association between the (STAT5a+, ERα+) breast cancer subgroup and the menopausal status. The same was found for the different subgroups. In addition, the different subgroups are strongly associated with high tumor grades (SBRII and SBRIII) but there is no significant association with node infiltration. The (STAT5a + ERα+) and (STAT5a + PR+) are associated with a proliferation index > 14% (P = 0.05 and P = 0.006, respectively). All subgroups are associated with a negative status of Her2.
 Click to view | Table 3. Coexpression of STAT5a With ERα, ERβ and PR and the Relationship With Clinical and Molecular Settings |
The STAT5a mRNA level in (STAT5a + ERα+), (STAT5a + ERβ+) and (STAT5a + PR+) breast cancer subgroups according to clinical and molecular information
Analysis of the STAT5a mRNA level, according to the clinical and molecular setting in the (STAT5a + ERα+) and (STAT5a + ERβ+) breast cancer subgroups, shows the absence of any significant differences (Fig. 3a, b). Inversely, in the (STAT5a + PR+) subgroup, the STAT5a mRNA level increases for the postmenopausal status (P = 0.02) (Fig. 3c). In addition, a high STAT5a mRNA level is associated with node infiltration (P = 0.03) (Fig. 3c).
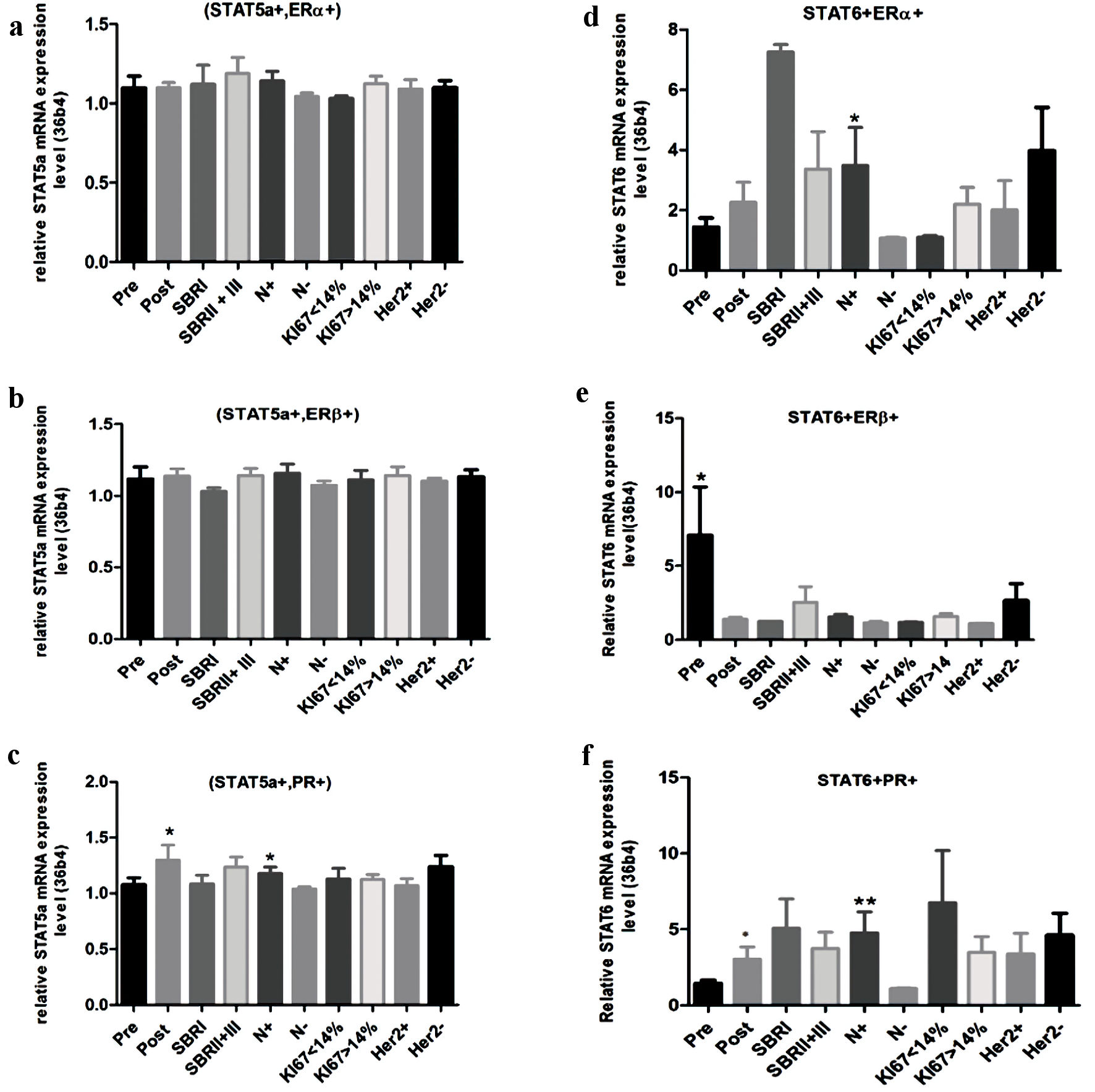 Click for large image | Figure 3. Relative mRNA expression level of STAT5a and STAT6 according to clinical and molecular information. (a) STAT5a mRNA expression level in (STAT5a+, ERα+) subgroup. (b) STAT5a mRNA expression level in (STAT5a+, ERβ+) subgroups. (c) STAT5a mRNA expression level in (STAT5a+, PR+) subgroups. (d) STAT6 mRNA expression level in (STAT6+, ERα+) subgroup. (e) STAT6 mRNA expression level in (STAT6+, ERβ+) subgroup. (f) STAT6 mRNA expression level in (STAT6+, PR+) subgroup. STAT: signal transducers and activators of transcription; ER: estrogen receptor; PR: progesterone receptor; SBR: Scarff-Bloom-Richardson. |
Relationship between (STAT6 + ERα+), (STAT6 + ERβ+) and (STAT6 + PR+) breast cancer subgroups and clinical settings
As shown in Table 4, there is the absence of a significant link between all subgroups and the menopausal status. Inversely, all subgroups are significantly associated with a high tumor grade. In addition, the coexpression of STAT6 with ERβ (but not with ERα) is strongly associated with node infiltration (P = 0.001). Further, (STAT6 + PR+) is significantly associated with infiltration nodes (metastasis) (P = 0.03). A significant association of (STAT6 + ERα+) and (STAT6 + PR+) subgroups with a proliferation index > 14% (P = 0.03 and P = 0.006, respectively) and a strong negative association of all subgroups with HER2/status have been found.
 Click to view | Table 4. Coexpression of STAT6 With ERα, ERβ and PR and the Relationship With Clinical and Molecular Settings |
The STAT6 mRNA level in (STAT5a + ERα+), (STAT5a + ERβ+) and (STAT5a + PR+) breast cancer subgroups according to clinical and molecular information
When STAT6 is coexpressed with ERα, the increase of the STAT6 mRNA level is significantly associated with node infiltration (P = 0.02) (Fig. 3d). In the (STAT6 + ERβ+) subgroup, the STAT6 mRNA level increases in the postmenopausal status (P = 0.01) (Fig. 3e). Finally, in the (STAT6 + PR+) breast cancer subgroup, increasing the STAT6 mRNA level was significantly associated with the postmenopausal status and with node infiltration (P = 0.05 and P= 0.001, respectively) (Fig. 3f).
| Discussion | ▴Top |
The present study shows that STAT5a and STAT6 are strongly expressed in malign breast tumors and can promote proliferation and metastasis in ERs and PR-positive breast cancer. In addition, positive STAT5a and STAT6 predict the response to endocrine therapy. We have shown here that STAT5a and STAT6 were strongly overexpressed in malign breast tumors (87.5% and 78.12%, respectively) (Table 1). Our results agree with previous studies [8, 9]. There is no relationship between the STAT5a and STAT6 expressions and the menopausal status; however, considering the relative amount of mRNA, the STAT5a expression was found to be higher in the postmenopausal status compared to premenopausal breast tumors. Inversely, the mRNA expression level of STAT6 was much higher (three-fold) in premenopausal patients. Our finding shows a significant association of the STAT5a and STAT6 expressions with the histological tumor grade. This result is concordant with the findings of Yamashita et al who demonstrated that the STAT5 expression was strongly correlated with the histological grade [19]. No links have been found between the STAT5a expression and lymph node metastasis. In this context, Wagner et al showed absence of a relationship between the STAT5a expression the menopausal status, the tumor size and metastasis [20]. On the contrary, the STAT6 expression is associated with node infiltration. This result is in agreement with a previous study that showed the association of STAT6 with node infiltration and distant metastasis [12, 21]. The analysis of the relative mRNA expressions of STAT5a and STAT6 shows the absence of a significant difference between node-positive and node-negative breast cancer. Inversely to previous data, our results reveal that STAT5a and STAT6 are both strongly associated with the tumor size. In addition, our results demonstrate a strongly negative association between the STAT5a and STAT6 expressions and the HER2/neu status. Consequently, STAT5 and STAT6 can predict the sensitivity to hormonal therapy. These results are in agreement with previous reports [19]. In addition, the present study reveals that there is no association between the STAT5a and STAT6 expressions and the proliferation index. This finding is concordant with the findings of Walker et al [22], which showed that the coactivation of STAT5 and STAT3 decrease cell proliferation in breast cancer, and with findings of Yu et al [23]. This showed that STAT5 negatively regulates cell proliferation by inducing cell cycle inhibitor genes.
Breast cancer is highly dependent on hormone receptors such as ERα, ERβ and PR, which play a crucial role in the initiation and progression of the disease [14-16, 24]. The analysis of the STAT5a and STAT6 expressions compared to the hormonal receptor expressions revealed a significant positive association of STAT5a and STAT6 with ERα and PR. These results are in agreement with previous studies which revealed an association of the STAT5 expression with the ER and PR positive statuses [19]. In addition, hyper-activation of STAT5a is associated with an increase of ERα [20]. The same is true for STAT6 which is expressed in ER+ breast cancer cell lines MCF-7 and ZR-75-1 [25].
The analysis of the STAT5a and the hormonal receptor (ERα, ERβ and PR) coexpressions compared to the clinical and molecular settings shows no association with the menopausal status, whereas there are significant associations of all breast cancer subgroups with the tumor grade. In addition, the (STAT5a + ERα+) and (STAT5a + PR+) breast cancer subgroups are associated with a high proliferation index. Consequently, STAT5a can interact with ERα and PR to induce tumor progression and cancer cell proliferation. Our finding is in agreement with previous studies which showed that STAT5a may be activated by several signaling pathways such as the estrogen one [26]. In fact, activation of STAT5 by estrogen pathways through c-Src can promote cell proliferation and survival in breast cancer [26]. In fact, STAT5a regulates the gene expression that promotes cell proliferation and survival, such as a heat shock protein 90-A (HSP90A) and cyclin D1 [27]. The expression and activities of STAT5 are also upregulated by PR [28]. In addition, the PR - STAT5 interaction can regulate gene and protein expressions [29]. Furthermore, the coexpression of STAT5a with ERs and PR is associated with a negative status of Her2/neu. In fact, the present study shows a strongly negative correlation between the (STAT5a+; ERβ+) and (STAT5a + PR+) breast cancer subgroups and the HER2/neu expression. A previous study showed links between Her2 downregulation and the sensitivity of endocrine therapy [30]. Consequently, the coexpression of STAT5a with ERs (especially ERβ) and PRs may be an indicator of endocrine sensitivity.
The same as it occurs with STAT5a, the coexpression of STAT6 with hormonal receptors (ERα, ERβ and PR) is not associated with the menopausal status, while all breast cancer subgroups ((ERα+, STAT6+) (ERβ+, STAT6+) (PR+, STAT6+)) are significantly associated with a high tumor grade (SBRII+ SBRIII). Moreover, the results of the present study demonstrate that an increase of the STAT6 mRNA expression is associated with infiltration nodes in (STAT6+, ERα+) and (STAT6+, PR+) breast cancer subgroups (Fig. 3b). In addition, the coexpression of STAT6 with ERβ and PR is strongly associated with node infiltration (Table 4). Our findings are concordant with pervious data that showed that STAT6 can affect tumor growth and metastatic niche formation [4-12, 21]. Furthermore, the association of the (STAT6+, ERβ+) and (STAT6+, PR+) breast cancer subgroups with infiltration nodes and the (STAT6+, ERα+) and (STAT6+, PR+) breast cancer subgroups with a high proliferation index shows the possibility of crosstalk between STAT6 and ERs and PR. Finally, the results show a strongly negative association of all breast cancer subgroups with the HER2/neu expression. This may be an indication of the sensitivity to hormonal therapy. Indeed, such as with STAT5a, the coexpression of STAT6 with ERs and PR may be a good indicator for endocrine therapy in association with the downregulation of HER2.
In conclusion, the results of the present study demonstrate that the STAT5a and STAT6 expressions were retained in the majority of the breast cancer cases. Often, STAT5a and STAT6 were coexpressed with ERs (ERα and ERβ) and PR. The coexpression of STAT5a and STAT6 with ERs and PR is associated with a high tumor grade, metastasis, and a high proliferation index. In fact, the (STAT5a+ ERα+), (STAT5a + PR+), (STAT6 + ERα+) and (STAT6 + PR+) breast cancer subgroups were associated with a high proliferation index while the (STAT6+ERβ+) and (STAT6+PR+) breast cancer subgroups were associated with metastasis. This finding means that STAT5a and STAT6 can interact with ERα, ERβ and PR and lead to cell proliferation and metastasis in malignant breast tumors.
Acknowledgments
The authors would like to thank the doctors and the technicians in the Surgical and Immuno-Histo-Cytology Departments of Salah Azaiez Institute.
Financial Disclosure
The present study was funded by The IMEC Unit, The Sciences Faculty of Bizerte, The University of Carthage and The Tunisian Ministry of Higher Education.
Conflict of Interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review.
Informed Consent
All the informed consents for publication were obtained.
Author Contributions
Mohamed Oueslati: study design, experimentation (immunohistochemistry and q-RT-PCR), results analysis and interpretation, writing of the manuscript. Ilhem Bettaieb determined the presence of malignant cells in the samples of breast cancer and did the analysis of immunohistochemistry results. Ridha Ben Younes contributed to the writing of the manuscript. Amor Gamoudi and Khaled Rahal determined the presence of malignant cells in the samples of breast cancer. Ridha Oueslati contributed to the results interpretation. All authors provided critical feedback and helped shape the research, analysis and manuscript.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
| References | ▴Top |
- Visvader JE, Lindeman GJ. Transcriptional regulators in mammary gland development and cancer. Int J Biochem Cell Biol. 2003;35(7):1034-1051.
doi - Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113(3):365-371.
doi pubmed - Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97-105.
doi pubmed - Haricharan S, Li Y. STAT signaling in mammary gland differentiation, cell survival and tumorigenesis. Mol Cell Endocrinol. 2014;382(1):560-569.
doi pubmed - Vafaizadeh V, Klemmt P, Brendel C, Weber K, Doebele C, Britt K, Grez M, et al. Mammary epithelial reconstitution with gene-modified stem cells assigns roles to Stat5 in luminal alveolar cell fate decisions, differentiation, involution, and mammary tumor formation. Stem Cells. 2010;28(5):928-938.
doi pubmed - Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11(2):179-186.
doi pubmed - Bertucci PY, Quaglino A, Pozzi AG, Kordon EC, Pecci A. Glucocorticoid-induced impairment of mammary gland involution is associated with STAT5 and STAT3 signaling modulation. Endocrinology. 2010;151(12):5730-5740.
doi pubmed - Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, Sauter G, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol. 2004;22(11):2053-2060.
doi pubmed - Cotarla I, Ren S, Zhang Y, Gehan E, Singh B, Furth PA. Stat5a is tyrosine phosphorylated and nuclear localized in a high proportion of human breast cancers. Int J Cancer. 2004;108(5):665-671.
doi pubmed - Iavnilovitch E, Cardiff RD, Groner B, Barash I. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int J Cancer. 2004;112(4):607-619.
doi pubmed - Khaled WT, Read EK, Nicholson SE, Baxter FO, Brennan AJ, Came PJ, Sprigg N, et al. The IL-4/IL-13/Stat6 signalling pathway promotes luminal mammary epithelial cell development. Development. 2007;134(15):2739-2750.
doi pubmed - Ostrand-Rosenberg S, Sinha P, Clements V, Dissanayake SI, Miller S, Davis C, Danna E. Signal transducer and activator of transcription 6 (Stat6) and CD1: inhibitors of immunosurveillance against primary tumors and metastatic disease. Cancer Immunol Immunother. 2004;53(2):86-91.
doi pubmed - Brisken C, O'Malley B. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2010;2(12):a003178.
doi pubmed - Skliris GP, Leygue E, Curtis-Snell L, Watson PH, Murphy LC. Expression of oestrogen receptor-beta in oestrogen receptor-alpha negative human breast tumours. Br J Cancer. 2006;95(5):616-626.
doi pubmed - Hopp TA, Weiss HL, Hilsenbeck SG, Cui Y, Allred DC, Horwitz KB, Fuqua SA. Breast cancer patients with progesterone receptor PR-A-rich tumors have poorer disease-free survival rates. Clin Cancer Res. 2004;10(8):2751-2760.
doi pubmed - Punglia RS, Kuntz KM, Winer EP, Weeks JC, Burstein HJ. The impact of tumor progesterone receptor status on optimal adjuvant endocrine therapy for postmenopausal patients with early-stage breast cancer: a decision analysis. Cancer. 2006;106(12):2576-2582.
doi pubmed - Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-408.
doi pubmed - Oueslati M, Bittaieb I, Sassi N, Jemaa AB, Gamoudi A, Rahal K, Oueslati R. ERalpha and ERbeta co-expression: An indicator of aggressive tumors and hormonal sensitivity. Oncol Lett. 2017;14(2):1675-1682.
doi pubmed - Yamashita H, Nishio M, Ando Y, Zhang Z, Hamaguchi M, Mita K, Kobayashi S, et al. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr Relat Cancer. 2006;13(3):885-893.
doi pubmed - Wagner KU, Rui H. Jak2/Stat5 signaling in mammogenesis, breast cancer initiation and progression. J Mammary Gland Biol Neoplasia. 2008;13(1):93-103.
doi pubmed - Binnemars-Postma K, Bansal R, Storm G, Prakash J. Targeting the Stat6 pathway in tumor-associated macrophages reduces tumor growth and metastatic niche formation in breast cancer. FASEB J. 2018;32(2):969-978.
doi pubmed - Walker SR, Nelson EA, Zou L, Chaudhury M, Signoretti S, Richardson A, Frank DA. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009;7(6):966-976.
doi pubmed - Yu JH, Zhu BM, Wickre M, Riedlinger G, Chen W, Hosui A, Robinson GW, et al. The transcription factors signal transducer and activator of transcription 5A (STAT5A) and STAT5B negatively regulate cell proliferation through the activation of cyclin-dependent kinase inhibitor 2b (Cdkn2b) and Cdkn1a expression. Hepatology. 2010;52(5):1808-1818.
doi pubmed - Lin Z, Reierstad S, Huang CC, Bulun SE. Novel estrogen receptor-alpha binding sites and estradiol target genes identified by chromatin immunoprecipitation cloning in breast cancer. Cancer Res. 2007;67(10):5017-5024.
doi pubmed - Furth PA. STAT signaling in different breast cancer sub-types. Mol Cell Endocrinol. 2014;382(1):612-615.
doi pubmed - Furth PA, Nakles RE, Millman S, Diaz-Cruz ES, Cabrera MC. Signal transducer and activator of transcription 5 as a key signaling pathway in normal mammary gland developmental biology and breast cancer. Breast Cancer Res. 2011;13(5):220.
doi pubmed - Rani A, Murphy JJ. STAT5 in Cancer and Immunity. J Interferon Cytokine Res. 2016;36(4):226-237.
doi pubmed - Richer JK, Lange CA, Manning NG, Owen G, Powell R, Horwitz KB. Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity. J Biol Chem. 1998;273(47):31317-31326.
doi pubmed - Cerliani JP, Guillardoy T, Giulianelli S, Vaque JP, Gutkind JS, Vanzulli SI, Martins R, et al. Interaction between FGFR-2, STAT5, and progesterone receptors in breast cancer. Cancer Res. 2011;71(10):3720-3731.
doi pubmed - Lindberg K, Helguero LA, Omoto Y, Gustafsson JA, Haldosen LA. Estrogen receptor beta represses Akt signaling in breast cancer cells via downregulation of HER2/HER3 and upregulation of PTEN: implications for tamoxifen sensitivity. Breast Cancer Res. 2011;13(2):R43.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Clinical Medicine Research is published by Elmer Press Inc.


