| Journal of Clinical Medicine Research, ISSN 1918-3003 print, 1918-3011 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Clin Med Res and Elmer Press Inc |
| Journal website http://www.jocmr.org |
Original Article
Volume 11, Number 2, February 2019, pages 98-105
Cytoskeletal Tropomyosin as a Biomarker in Clostridium difficile Infection
Elena A. Usachevaa, b, d, Lance R. Petersona, b, Katherine Mendozaa, Donna M. Schoraa, M. Moazzem Hossainc, Jian-Ping Jinc
aInfectious Disease Research, NorthShore University HealthSystem, 2650 Ridge Ave., Evanston, IL 60201, USA
bUniversity of Chicago Pritzker School of Medicine, Chicago, IL, USA
cDepartment of Physiology, Wayne State University School of Medicine, Detroit, MI 48201, USA
dCorresponding Author: Elena A. Usacheva, NorthShore University HealthSystem, 2650 Ridge Ave., Walgreen SB #525, Evanston, IL 60201, USA
Manuscript submitted November 18, 2018, accepted December 24, 2018
Short title: Biomarker in C. difficile Infection
doi: https://doi.org/10.14740/jocmr3696
| Abstract | ▴Top |
Background: Current diagnostics of Clostridium difficile infection (CDI) heavily relies on detection of the disease-causing organism. The objective of this study was to investigate a cytoskeletal protein, tropomyosin (Tpm), as a CDI biomarker.
Methods: Fecal Tpm was tested by monoclonal antibodies (mAbs) in a 12-month prospective study. Remnant diarrheal clinical specimens and relevant clinical data were collected. The CDI positive (CDI+, n = 230) and CDI negative (CDI-, n = 228) groups were composed of samples testing positive or negative by polymerase chain reaction (PCR) (Xpert® C. difficile/Epi, Cepheid), respectively. The other enteric pathogen (OEP) group (n = 52) was composed of specimens tested for the presence of other enteric pathogens or parasites by routine testing methods. Extracted fecal Tpm was detected by Western blot and the results were correlated with CDI based on clinical and microbiology laboratory data.
Results: A total of 510 stool specimens were tested. Tpm is not stable in stool, suggesting the utility of fresh specimens. In the CDI+ group, specificity and sensitivity of Tpm detection in correlation with a CDI were 93.2% and 53.7%, respectively, when only “true CDI” and “not CDI” were analyzed (110 samples). For CDI+ samples, 23% did not satisfy CDI clinical signs. Tpm positives in the CDI- group (8.3%) had inflammatory bowel diseases.
Conclusion: Tpm has a potential role as a CDI biomarker in combination with C. difficile PCR and an appropriate clinical evaluation. However, non-muscle Tpm, as a biomarker for CDI, suffers from a low sensitivity in our study. Therefore further investigation using larger cohorts is needed.
Keywords: Clostridium difficile infection; Biomarker; Tropomyosin; Diagnostics
| Introduction | ▴Top |
Clostridium difficile infection (CDI) is a significant health concern worldwide and is recognized as the most frequent etiologic agent of healthcare-associated infectious diarrhea in hospitalized adult patients [1]. For the last two decades, CDI has re-emerged in healthcare facilities with nearly a 10-fold increase in mortality [2]. In the USA alone, C. difficile was associated with approximately 29,000 deaths in 2011 and exerted significant impact on the length of hospital stay and cost, estimated at more than $850 million in excess of healthcare expense annually [3-5]. The epidemiology of the disease is changing with increasing community-acquired CDI, a population without traditional risk factors [6-8]. C. difficile is increasingly resistant to a broad range of antibiotics demonstrating an emerging pattern of resistance to available treatment and recurrence after an initial episode [9].
There is no specific diagnostic test for this disease. Laboratory testing available for CDI identification includes sigmoidoscopy and colonoscopy, toxigenic culture (TC), cell cytotoxicity assay (CCTA), enzyme immunoassay (EIA), glutamate dehydrogenase (GDH) EIA, as well as very sensitive real-time polymerase chain reaction (rtPCR) and loop-mediated isothermal amplification assay (LAMP) detecting DNA of C. difficile toxin(s) [10]. The current diagnostic strategy relies on combination of clinical signs and symptoms (frequency of diarrhea, antibiotic exposure, and white blood cell count elevation) with a positive diagnostic test for toxigenic C. difficile in stool. However, C. difficile PCR tests, TC, and CCTA only detect the presence of a disease-causing organism or its toxins and when positive they are not diagnostic of clinical infection [10]. This organism can colonize 10-fold more patients asymptomatically than those actually develop infection [11].
C. difficile pathogenicity is primarily dependent on the presence of one or both diarrhea-producing toxins named toxin A and toxin B (the latter is always essential for the pathogenicity). The molecular mechanism of the toxin action is an enzymatic inactivation of intracellular Rho GTPase leading to the depolymerization of actin filaments, cytoskeletal disruption and subsequent cell death [12-14]. Cell adhesion is dependent on the function of actin cytoskeleton [15], and the effects of C. difficile toxin result in accelerated dissociation of colonic epithelial cells. This, in turn, leads to significant loss of the intestinal epithelial barrier and stimulates an inflammatory cascade causing tissue damage, diarrhea and pseudomembranous colitis [16, 17]. We hypothesize that this mechanism permits release of intracellular protein(s) that can be used as a measure of toxin B effect on human colonic tissue in C. difficile infection.
Tropomyosin (Tpm) is a protein family associated with the stabilization and regulation of the actin cytoskeleton [18]. It is expressed in eukaryotes including animals and fungi, but is not documented in plants, protists or prokaryotes. Tpm isoforms show a wide range of abundance in different tissues such as brain, heart, kidney, liver, lung, stomach and spleen [19, 20]. Several isoforms of Tpm constitute a family of actin-binding proteins, important components of both muscle contractile machinery and non-muscle cell cytoskeleton. This protein is critical for cytoskeletal function in non-muscle cells by stabilizing actin filaments [21]. Tpm is a highly soluble about 33 kDa protein with an isoelectric point of about 4.6 [22] and it is stable during biochemical isolation, rendering it a candidate for use as a biomarker to detect colon epithelial damage and indicate the release of cytoskeletal proteins in CDI. Mirza et al demonstrated that human Tpm isoform 5 (hTpm5), but not other isoforms, are expressed in the epithelium of the colon [23], suggesting a specific target for such detection.
The aim of the present study is to investigate the feasibility of fecal non-muscle cytoskeletal Tpm as a host response-specific biomarker for active infection of C. difficile.
| Material and Methods | ▴Top |
Experimental design
Tpm utility for recognizing patients with diarrhea due to CDI was investigated by testing toxigenic C. difficile negative and positive stool specimens (determined by the Xpert® C. difficile/Epi, Cepheid, Sunnyvale, CA): “CDI-” group and “CDI+” group, respectively. Tpm was also tested in stool specimens from patients with diarrheal illness other than CDI such as functional bowel disorders and infection with other enteric pathogens or parasites (other enteric pathogen group, OEP). These specimens usually are not tested for C. difficile toxin B DNA.
The presence of fecal Tpm was demonstrated by detection with monoclonal antibodies (mAbs) against human non-muscle tropomyosin (hTpm). Central to this investigation was the correlation of Tpm detection (host cellular response) with CDI clinical manifestations and symptoms supplemented with microbiology data referred as the “reference approach” (Fig. 1).
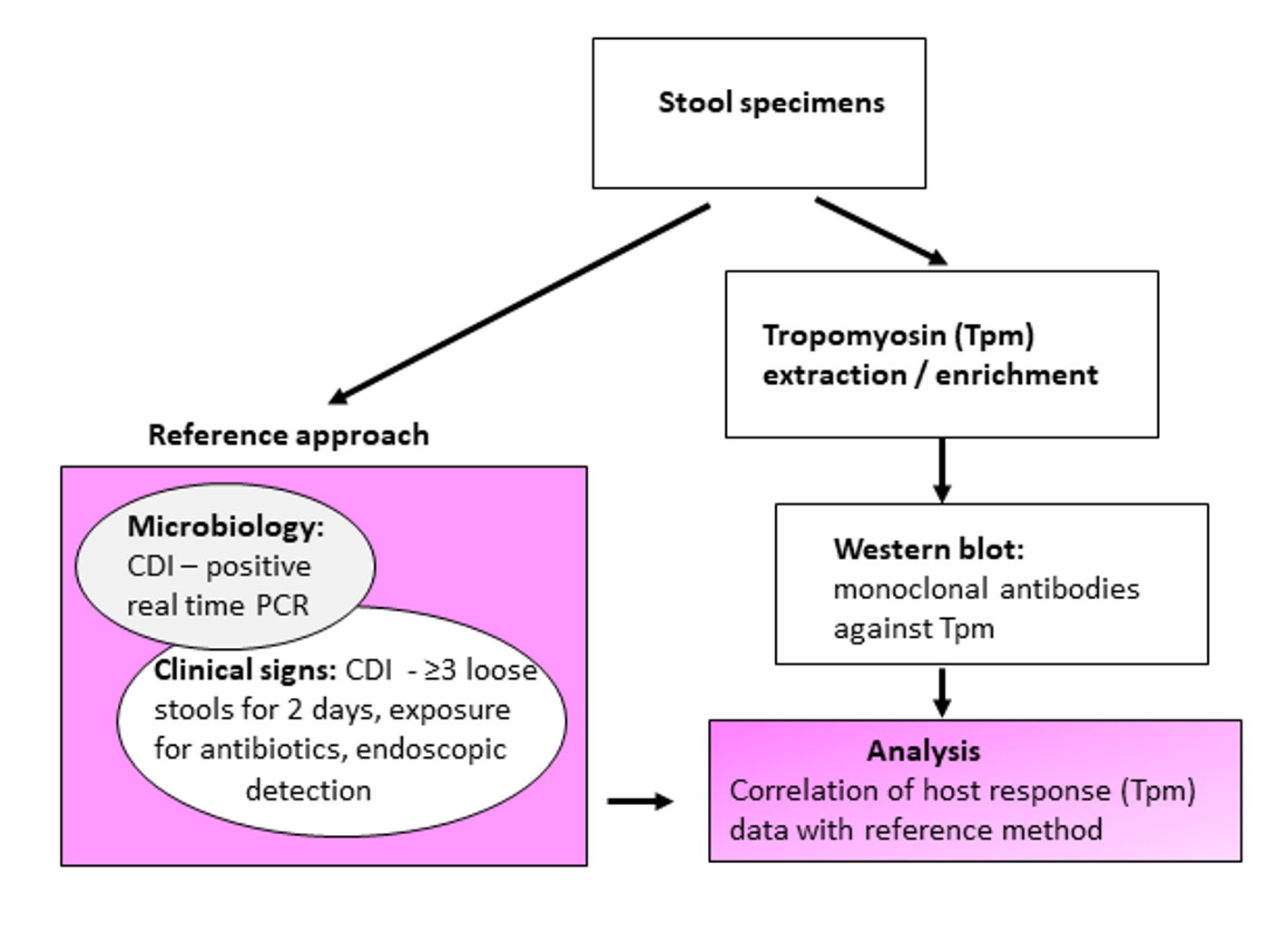 Click for large image | Figure 1. Schematic experimental design and evaluation of tropomyosin (Tpm) as a Clostridium difficile infection (CDI) biomarker. |
The definition used for the reference true CDI positive specimens satisfied two key Tpms: 1) Microbiology (detection of C. difficile toxin B gene by rtPCR, the Xpert® C. difficile/Epi); and 2) Clinical symptoms (at least three loose stools during 2 consecutive days and prior antibiotic exposure, or visual endoscopic detection of pseudomembranes). Any deviation from these criteria implied a CDI negative result by the reference approach. Specimens tested positive by the Xpert® C. difficile/Epi but with undetermined diagnosis/incomplete clinical symptoms’ data were considered as equivocal and excluded from analysis.
Patients and specimens
The study was approved by Institutional Review Board (IRB: EH14-229). Specimens used for the study were prospectively collected remnant de-identified diarrheal stool specimens submitted to NorthShore University HealthSystem’s Clinical Microbiology Laboratory for routine testing (Fig. 2). Collected specimen information comprised of C. difficile PCR result, detection of enteric pathogens other than C. difficile, or parasites, number of stools on the collection day and the prior day, antibiotic consumption within 6 months, and gastrointestinal diseases (e.g. ulcerative colitis or Crohn’s disease). Stool selection criteria for Tpm testing were defined as loose or watery consistency up to 3 days from specimen collection. Additionally, five assigned study participants (from a cohort of 510 samples: the Result section) provided written informed consent before sampling.
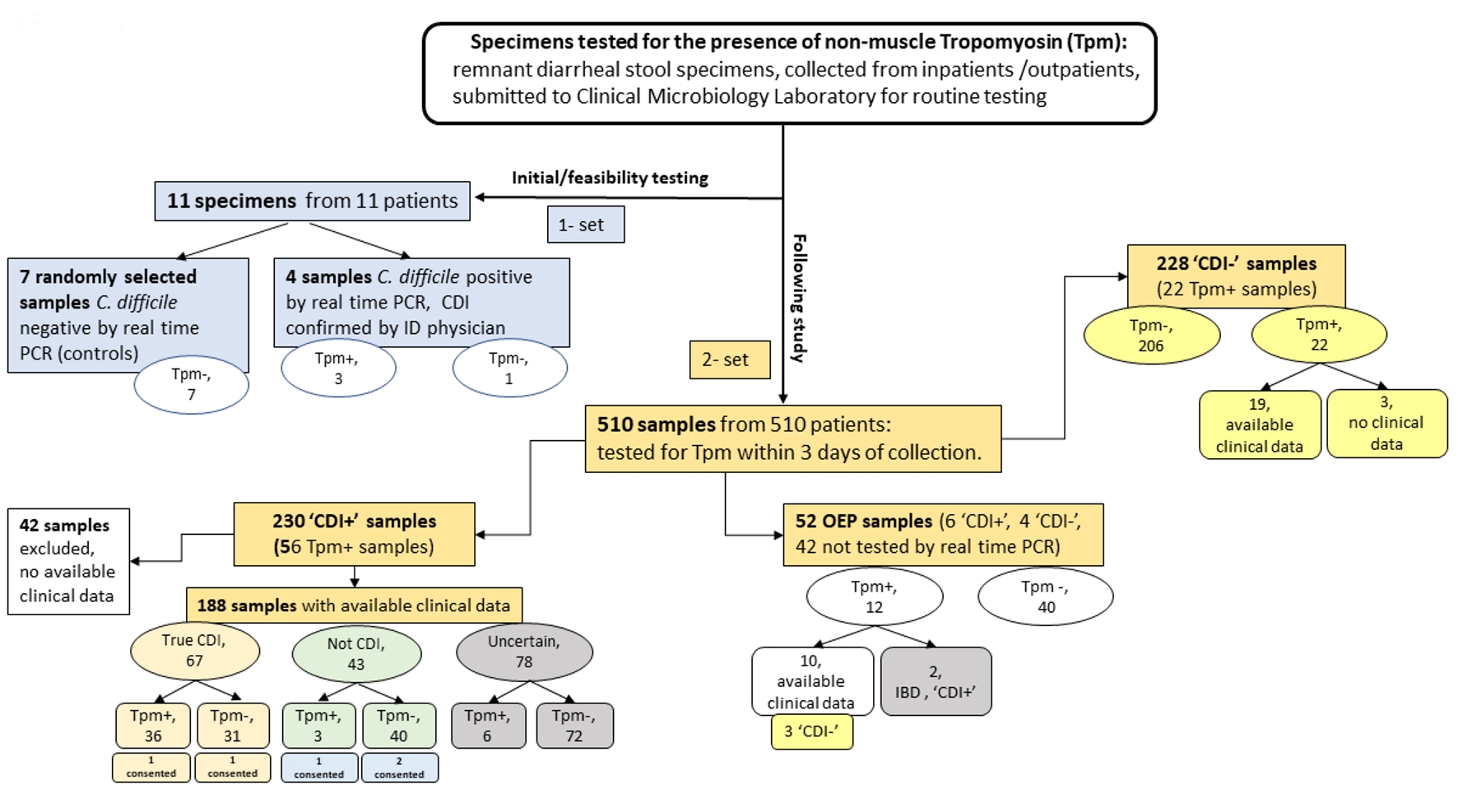 Click for large image | Figure 2. Distribution of samples tested for tropomyosin. CDI: C. difficile infection; ID: an infectious disease physician; OEP: other enteric pathogens; “Tpm-”: tropomyosin is not detected; “Tpm+”: tropomyosin is detected; “CDI+”: specimens tested positive by rtPCR (the Xpert® C. difficile/Epi, Cepheid); “CDI-”: specimens tested negative by rtPCR (the Xpert® C. difficile/Epi, Cepheid); “true CDI”: compatible with CDI according to the reference approach (microbiology and clinical evaluation); “not CDI”: deviated from the reference approach; “Uncertain”: specimens tested positive by PCR but the diagnosis is inconclusive; “Consented”: sample was collected from consented patient. In the groups of “CDI-” and “OEP”, clinical signs were evaluated only for “Tpm+” samples. |
Human colon tissues, used for assessment of hTpm isoforms in colon cells, were discarded surgical samples of colon cancer and normal tissues processed immediately after the surgical procedure. The tissue (1 × 1 cm) was rinsed three times with saline to remove colon contents and mucus, dried on paper, and stored at -80 °C. The time between surgical procedure and freeze was no longer than 15 min. The tissue included all layers of the colon wall. Total proteins were extracted from total colon wall tissue. Briefly, the tissue was cut into small pieces, high-speed homogenizer was used to release proteins and the mixture was heated at 80 °C. The supernatant was further clarified by centrifugation at 14,000 g, and the clean supernatant was then subjected to further procedures.
Development of mAbs
Human non-muscle Tpm was expressed from cloned cDNA by transformation of E. coli cultures with expression plasmids pKK233-2 (Pharmacia, NJ, USA). Briefly, the entire coding region of Tpm isoform cDNA was obtained from pSMT-10 by NcoI digestion [24]. The 990 bp NcoI fragment was subcloned into the expression vector pKK233-2. Modified from Novy et al [24], recombinant human non-muscle Tpm was purified using ion-exchange and gel filtration chromatographic columns, dialyzed to remove salts and lyophilized.
The study was approved by the Institutional Animal Care and Use Committee (IACUC) in compliance with the Animal Welfare Act and Regulations. As described previously [25], purified human non-muscle Tpm was used to immunize Balb/c mice. Spleen cells of a short-term immunized mouse were harvested for fusion with SP2/0 mouse myeloma cells. Hybridomas were selected with HAT media, screened for anti-Tpm mAb production using indirect enzyme-linked immunosorbent assay (ELISA) and Western blotting against the immunogen, and subcloned three or more times to establish stable cell lines. After determining immunoglobulin isotype using a sandwich ELISA kit (BD Biosciences, San Jose, CA), the anti-human non-muscle Tpm mAb was made in the forms of hybridoma culture supernatant and mouse ascites fluids.
Stool specimen processing and detection
For this study a two-step approach leading to Tpm detection was developed: crude extraction from stool and detection with Tpm specific mAbs. Two grams of stool samples were brought to 5 mL volume with phosphate buffer saline (PBS), pH 7.8 containing 1 mg/mL phenylmethanesulfonyl fluoride (PMSF) in 15-mL centrifuge tube, boiled for 20 min, with mixing every 5 min, cooled on ice for 30 min with mixing, and spun at 3,000 g at 4 °C for 20 min to remove most of the insoluble stool contents. The supernatant was further clarified by centrifugation in a microcentrifuge at 14,000 g at 4 °C for 10 min. The clean supernatant of 1.4 mL was then subjected to isoelectric point precipitation to enrich Tpm. This was accomplished by adjusting the sample pH to 4.5 - 4.6 by adding 50 µL 3M sodium acetate, pH 4.2 and incubation at 4 °C for 10 min with rotation, and centrifugation at 14,000 g for 30 min at 4 °C to collect Tpm in the pellet. The pellets were either dissolved in 20 µL 1 × Laemmli SDS-PAGE sample buffer (Bio-Rad Laboratories, Inc., Des Plaines, IL), or stored at -80 °C. The remaining supernatant was stored at -80 °C as well.
Fifteen microliters of the pellet SDS-PAGE sample were heated at 80 °C for 5 min and loaded on 12% SDS-PAGE (Mini-Protean TGX, Bio-Rad Laboratories, Inc., Des Plaines, IL). The protein bands resolved were transferred to 0.45 µm nitrocellulose membrane (Bio-Rad Laboratories, Inc., Des Plaines, IL) using a wet Western blotting transfer apparatus (Mini Trans-Blot, Bio-Rad Laboratories, Inc., Des Plaines, IL). Antibody reaction was performed on iBind Western System (Life technology, NY, NY). Goat Anti-Mouse IgG-AP was used as secondary antibody in combination with a colorimetric immunodetection kit (Bio-Rad Laboratories, Inc., Des Plaines, IL).
Statistical analysis
For sample size estimation we hypothesized that Tpm diagnostic method will have a sensitivity and specificity of 0.95. A total sample size of 300 (which includes 150 subjects with CDI and 150 non-CDI subjects) achieves 90% power to detect difference in sensitivity between 0.8 and 0.95 using a two-sided binomial test and 90% power to detect difference in specificity between 0.8 and 0.95 using a two-sided binomial test with a significance level of 0.05. The prevalence of the disease in the tested samples was structured to be 0.5. Thus, to demonstrate good diagnostic ability of host response detection, we needed at least 150 specimens that will result as positive by “reference method” (positive by Xpert® C. difficile/Epi assay and CDI clinical signs).
All duplicated samples were removed for data analysis. Fisher’s exact test was used for comparative analysis of Tpm detection in “CDI+” group with correlation to significant CDI clinical symptoms. P value of < 0.05 was considered statistically significant. The specificity and sensitivity of our Tpm detection test were determined using standard formulas for specimens defined as “true CDI” and “not CDI”. Since the prevalence of the disease in the tested samples did not reflect the true disease prevalence, positive and negative predictive values (PPVs and NPVs) were not calculated. Tpm detection for “positive” and “negative” cases was summarized as frequency and percentages (n; %; Tables 1 and 2).
 Click to view | Table 1. Tpm Detection and Distribution Within “CDI+” Group |
 Click to view | Table 2. Tpm Detection and Distribution Within “CDI+” Group in Inpatients |
| Results | ▴Top |
Feasibility of Tpm detection on clinical samples
A convenience sample of 11 prospectively collected stool specimens were analyzed in parallel by SDS-PAGE and Western blot using a combination of anti-Tpm mAbs (Fig. 3). Seven patients were tested negative for C. difficile and four were positive. All four patients with positive tests for C. difficile toxin gene (the Xpert® C. difficile/Epi, Cepheid) were evaluated by an infectious disease physician to assure that all had disease compatible with CDI. Samples 1 - 7 were randomly selected from diarrheal stool samples submitted to the laboratory that were negative for toxigenic C. difficile. Patient 8 did not show a detectable Tpm amount in the crude stool sample.
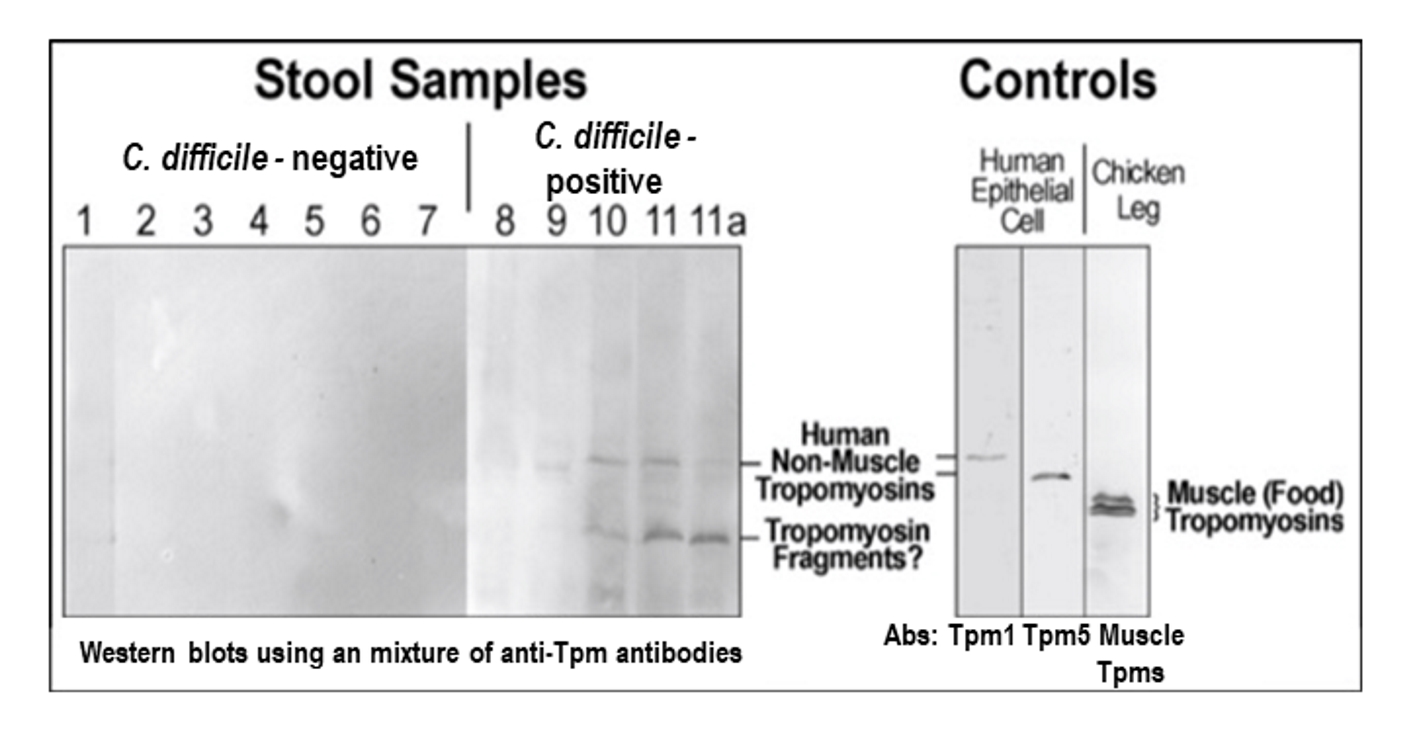 Click for large image | Figure 3. Detection of non-muscle tropomyison (Tpm) in stool samples. Samples #8-11 were collected from CDI patients. Samples #9-11 are positive for Tpm (upper bend). #11a: after the treatment, upper band is disappeared. The mixture of anti-Tpm antibodies (Abs) is comprised of monoclonal antibodies for Tpm1, Tpm 5, and chicken leg muscle Tpm. To control a loading amount of proteins an equal amount of stool starting material was always used for Tpm crude extraction. |
Protein extracts from HT29 human colon epithelial cell culture were used as a positive control and chicken leg muscle extracts were used as a negative control to exclude food (meat/muscle) protein contents. The results demonstrated a detection of Tpm in samples with positive C. difficile culture in three of four patients tested (#9-11). Sample #11a was retaken from patient #11 when the C. difficile infection had clinically resolved as documented by an infectious disease physician review, and the Tpm level decreased noticeably in this sample. The sample was not retested for C. difficile toxin DNA as test of cure for C. difficile is not recommended once the clinical disease has resolved.
The data indicated that the Tpm antibody mixture did not detect anything in all control patient samples (Fig. 3, #1-7), although individual muscle Tpm antibodies did detect chicken muscle Tpm at a different molecular weight in muscle control (the right panel, Fig. 3). Thus, it excluded food Tpm as a likely source of false positives. Secondly, the results suggested that Tpm levels increased considerably when patients had disease compatible with CDI.
Tpm stability in clinical stool samples
Four mAbs were generated for detection of Tpm and tested on three controls and seven well characterized clinical stool specimens. Only two of them were considered for further testing due to their Tpm specificity and robust signal (a band of about 33 kDa corresponding to the molecular weight of Tpm).
Originally, the Tpm extraction from each stool sample was performed within 7 days from collection. To investigate Tpm stability in clinical specimens, the protein contents were extracted twice from aliquots of the same sample stored at refrigerator (4 - 10 °C). The first Tpm extraction was performed within 1 - 3 days from stool collection, and the second extraction was done in 7 days. Of the 10 samples that were Tpm positive with the first extraction, three failed to detect Tpm when extracted at 7 days. This observation suggests that Tpm may not be stable in clinical stool specimens and ideally should be extracted within 1 day from collection. Consistent with this observation, stool samples collected 2.5 months apart from the same patient were tested Tpm negative when extracted on the third day from collection but detected positive when extracted the day after collection.
Of note, Tpm was successfully extracted from frozen samples (-80 °C) stored up to 6 months.
Detection of fecal Tpm and correlation with CDI
From December 2016 to November 2017, we prospectively collected 510 stool specimens (Fig. 2) from different patients for Tpm interrogation (Table 3). Two hundred thirty “CDI+” samples were collected of which 56 resulted Tpm positive. However, 42 specimens in “CDI+” did not have sufficient clinical data therefore reducing the numbers to 188 specimens available for analysis (Table 3). In the “CDI+” group, 24% of stool specimens were tested Tpm positive (45/188) (Table 1). Of them 80% (36/45 samples) correlated with true CDI, three cases were considered as inflammatory bowel disease (IBD) or viral infection rather than CDI, and etiology of remaining six cases was hard to determine based on clinical data (samples with uncertain diagnosis). In overall “CDI+” group, Tpm was not detected in 76% (143/188) stool specimens. The diagnosis was not defined in majority of these specimens (n = 72) and in 21.7% (31/143 samples) true CDI was not correlated with Tpm detection. Of note, 23% (43/188 samples) PCR-tested positives did not satisfy true CDI criteria (93% of these tested as Tpm negative).
 Click to view | Table 3. Clinical Stool Specimens Tested for Tropomyosin (Tpm) |
Focused only on “true CDI” and “not CDI” groups (total 110 samples) by eliminating samples with uncertain diagnosis (Table 1), the specificity and sensitivity of Tpm detection in correlation with CDI diagnosis was estimated as 93.2% and 53.7%, respectively, with the P value < 0.0001.
Stratified by inpatient criteria, 26% (49/188 samples) were collected from inpatients (Table 2). Of these inpatients, 82% (9 /11 samples) in the Tpm positive category were true CDI. However, no correlation with CDI diagnosis was demonstrated in Tpm negatives. By eliminating samples with uncertain CDI diagnosis for inpatients, the 32 specimens demonstrated 93% specificity and 50% sensitivity with P value of 0.02.
Five patients were consented and interviewed in regards to their CDI symptoms. Two patients satisfied the true CDI criteria and were determined as “true CDI” patients while the others had IBD and acquired C. difficile colonization. However, out of the two CDI patients only one tested Tpm positive.
Two hundred twenty-eight “CDI-” samples were collected (Table 3). In this group only 22 samples were tested Tpm positive with majority provided by patients with IBDs (19/228; 8.3%): Crohn’s (30%; 7/19) and colitis (52%; 12/19). However, we should point out that not all samples tested from patients with IBD correlate with Tpm detection.
The OEP group was comprised of 52 stool specimens positive for one or more pathogens such as Campylobacter jejuni, Campylobacter coli, Aeromonas veronii, Shigella boydii, Salmonella enteritidis, shigatoxigenic E.coli, or Blastocystis hominis. Nineteen percent (10/52 samples) were tested Tpm positive (Table 3) and 77% (40 Tpm negatives /52 samples) Tpm negative, with P = 0.05. All of the Tpm positives were not correlated with IBD. Of note, Giardia lamblia sample was tested Tpm negative.
| Discussion | ▴Top |
In our prospective clinical investigation, we tested cytoskeletal protein, Tpm, as a biomarker for CDI diagnostics in conjunction with a rtPCR-based assay (Xpert® C. difficile/Epi). The selection of non-muscle cytoskeletal Tpm as the preferred indicator to monitor actin cytoskeleton integrity in C. difficile-affected colonic epithelial cells was justified by: 1) Its abundance in colon epithelial cells (0.2% of the cellular proteins); and 2) Heat resistance to allow rapid and practical enrichment from stool. MAbs for recognition of non-muscle Tpm were generated and used for the protein detection in clinical stool specimens. We demonstrated that Tpm detection in samples positive by C. difficile PCR highly correlated with clinically relevant CDI with a specificity > 90%. However, we also demonstrated that cases of true CDI may lack fecal Tpm. This observation was corroborated in samples from two consented patients with confirmed clinically relevant CDI. Only one of them provided a Tpm positive result. This implies that non-muscle Tpm as a biomarker for CDI suffers from low sensitivity, but needs further investigation to study a larger size of samples. We anticipate that implementing antibodies with enhanced affinity to non-muscle Tpm and utility of fresh or better preserved specimens may improve Tpm detection sensitivity as a CDI biomarker. Additionally, while we did not observe clear quantitative correlation between Tpm detection and CDI severity (out of the study score), the use of improved antibodies on large size of specimens may elucidate Tpm potential as a biomarker for CDI severity.
Asymptomatic C. difficile colonization is the absence of symptoms with the prevalence range from 1.4% to 21% [26, 27]. Currently, widely implemented in clinical laboratories, the rtPCR assay for detection of C. difficile (usually toxin B) can lead to misclassification of some C. difficile carriers as CDI cases [28]. A few fecal biomarkers of inflammation have been investigated over the last decades. Some of these markers have been shown to be produced in response to C. difficile toxins. However, none of them have been demonstrated to be specific for CDI in the general population (Table 4). Our findings might indicate that patients testing positive for both C. difficile PCR and Tpm have true CDI.
 Click to view | Table 4. Summary of Fecal Inflammatory Biomarkers as Possible Predictors of CDI Disease* |
Interestingly, Tpm detection in the “CDI-” group was always observed in concordance with IBD, the majority colitis (ulcerative/collagenous/lymphocytic diverticulitis) and Crohn’s disease. This may reflect the fact that disruptions of the colon epithelia in IBD and CDI are similar. Therefore, it may complicate the results in patients testing positive for Tpm/C. difficile PCR/IBD, making the accurate diagnosis of CDI in these patients difficult.
While C. difficile is the most common infectious etiology of antibiotic-associated diarrhea (AAD), only 25% of all AAD cases are associated with CDI [29, 30]. Numerous other bacterial infectious agents have been implicated in AAD, including Clostridium perfringens, Staphylococcus aureus, and Klebsiella oxytoca [30]. Our clinical laboratory routine does not imply testing for these pathogens in diarrheal stools. However, a few representative C. difficile PCR positive stool samples were cultured for detection of C. difficile colonies. Some of them did not grow C. difficile while the others grew Clostidium spp. such as C. innocuum and C. buturicum (not shown; detected by MALDI-TOF MS, Brucker). This might be an indication of another etiology of diarrhea in the presence of C. difficile colonization.
In the OEP group all Tpm positives were tested positive for Campylobacter jejuni, Campylobacter coli, Aeromonas veronii, Shigella boydii, Salmonella enteritidis, shigatoxigenic E. coli, or Blastocystis hominis. However, none of these pathogens correlated with Tpm positivity in this group. Furthermore, the pathogens were detected in Tpm-negative samples as well. A small number in this cohort and wide range of detected pathogens suggest a need of more extended studies.
The main limitations of our study were limited clinical data for correlation with clinically relevant CDI, and the small sample size of inpatients. Focusing on inpatients, the intention was to take advantage of more complete data collected for this patient population. However, the four NorthShore hospitals (789 beds) were not able to provide sufficient number of inpatients during the study period. A small number of inpatient specimens were collected (Table 2). Among them “uncertain samples” comprised 34.7% (17/49 samples). This highlights the complexity of the disease signs and symptoms, and many confounders complicate the diagnosis. Thus, a high proportion of Tpm-negative inpatients in the “CDI+” group were also confounded with diverse types of cancer (undergoing chemotherapy) or colitis. Therefore, etiology of the diarrhea was unclear. For this small number of inpatients, Tpm detection specificity and sensitivity for clinically relevant CDI was determined as of 93% and 50%, respectively. The lack of proper CDI diagnostics highlights the fact that in this study we demonstrated that almost 23% of C. difficile PCR positives were misdiagnosed, detecting C. difficile colonization rather than clinical CDI.
Another limitation was that culture was not performed on all samples in the “CDI+” group. A larger sample size might have helped us find a stronger correlation between Tpm and CDI.
Future studies should be focused on a large number of patients, testing with antibodies recognizing multiple Tpm epitopes, and the investigation of mechanisms of colon epithelial Tpm release in response to C. difficile toxins.
Conclusions
One of the challenges of managing CDI is the initial diagnosis of the disease. To date, there is no single test that accurately and rapidly diagnoses CDI. Based on the mechanism of action of C. difficile toxins on human colonic tissue, we have attempted to propose and evaluate a novel biomarker for CDI. Our investigation highlights the fact that Tpm has a potential role as a biomarker of CDI when performed in combination with C. difficile PCR and an appropriate clinical evaluation.
Conflict of Interest
The authors declare that they do not have any conflict of interest.
Financial Support
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R21AI116659 to EAU and JPJ.
Author Contributions
All authors participated to review. All authors were involved in writing and revising the article prior to submission.
| References | ▴Top |
- Khanna S, Pardi DS. Clostridium difficile infection: management strategies for a difficult disease. Therap Adv Gastroenterol. 2014;7(2):72-86.
doi pubmed - Lessa FC, Gould CV, McDonald LC. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis. 2012;55(Suppl 2):S65-70.
doi pubmed - Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825-834.
doi pubmed - Schroeder LF, Robilotti E, Peterson LR, Banaei N, Dowdy DW. Economic evaluation of laboratory testing strategies for hospital-associated Clostridium difficile infection. J Clin Microbiol. 2014;52(2):489-496.
doi pubmed - Tabak YP, Zilberberg MD, Johannes RS, Sun X, McDonald LC. Attributable burden of hospital-onset Clostridium difficile infection: a propensity score matching study. Infect Control Hosp Epidemiol. 2013;34(6):588-596.
doi pubmed - Kuntz JL, Chrischilles EA, Pendergast JF, Herwaldt LA, Polgreen PM. Incidence of and risk factors for community-associated Clostridium difficile infection: a nested case-control study. BMC Infect Dis. 2011;11:194.
doi pubmed - Chitnis AS, Holzbauer SM, Belflower RM, Winston LG, Bamberg WM, Lyons C, Farley MM, et al. Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern Med. 2013;173(14):1359-1367.
doi pubmed - Ogielska M, Lanotte P, Le Brun C, Valentin AS, Garot D, Tellier AC, Halimi JM, et al. Emergence of community-acquired Clostridium difficile infection: the experience of a French hospital and review of the literature. Int J Infect Dis. 2015;37:36-41.
doi pubmed - Ng QX, Loke W, Foo NX, Mo Y, Yeo WS, Soh AYS. A systematic review of the use of rifaximin for Clostridium difficile infections. Anaerobe. 2018;55:35-39.
doi pubmed - Usacheva EA, Jin JP, Peterson LR. Host response to Clostridium difficile infection: Diagnostics and detection. J Glob Antimicrob Resist. 2016;7:93-101.
doi pubmed - Galdys AL, Nelson JS, Shutt KA, Schlackman JL, Pakstis DL, Pasculle AW, Marsh JW, et al. Prevalence and duration of asymptomatic Clostridium difficile carriage among healthy subjects in Pittsburgh, Pennsylvania. J Clin Microbiol. 2014;52(7):2406-2409.
doi pubmed - Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev. 2005;18(2):247-263.
doi pubmed - Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, Haslam DB, Goldenring JR, Lacy DB. Clostridium difficile Toxin B causes epithelial cell necrosis through an autoprocessing-independent mechanism. PLoS Pathog. 2012;8(12):e1003072.
doi pubmed - Rupnik M. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol Rev. 2008;32(3):541-555.
doi pubmed - Buda A, Pignatelli M. Cytoskeletal network in colon cancer: from genes to clinical application. Int J Biochem Cell Biol. 2004;36(5):759-765.
doi pubmed - Burnham CA, Carroll KC. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin Microbiol Rev. 2013;26(3):604-630.
doi pubmed - Burke KE, Lamont JT. Clostridium difficile infection: a worldwide disease. Gut Liver. 2014;8(1):1-6.
doi pubmed - Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil. 2015;36(2):147-153.
doi pubmed - Wang CL, Coluccio LM. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int Rev Cell Mol Biol. 2010;281:91-128.
doi - Schevzov G, Vrhovski B, Bryce NS, Elmir S, Qiu MR, O'Neill G M, Yang N, et al. Tissue-specific tropomyosin isoform composition. J Histochem Cytochem. 2005;53(5):557-570.
doi pubmed - Lin JJ, Eppinga RD, Warren KS, McCrae KR. Human tropomyosin isoforms in the regulation of cytoskeleton functions. Adv Exp Med Biol. 2008;644:201-222.
doi pubmed - Lin JJ, Warren KS, Wamboldt DD, Wang T, Lin JL. Tropomyosin isoforms in nonmuscle cells. Int Rev Cytol. 1997;170:1-38.
doi - Mirza ZK, Sastri B, Lin JJ, Amenta PS, Das KM. Autoimmunity against human tropomyosin isoforms in ulcerative colitis: localization of specific human tropomyosin isoforms in the intestine and extraintestinal organs. Inflamm Bowel Dis. 2006;12(11):1036-1043.
doi pubmed - Novy RE, Liu LF, Lin CS, Helfman DM, Lin JJ. Expression of smooth muscle and nonmuscle tropomyosins in Escherichia coli and characterization of bacterially produced tropomyosins. Biochim Biophys Acta. 1993;1162(3):255-265.
doi - Liu R, Hossain MM, Chen X, Jin JP. Mechanoregulation of SM22alpha/Transgelin. Biochemistry. 2017;56(41):5526-5538.
doi pubmed - Kong LY, Dendukuri N, Schiller I, Bourgault AM, Brassard P, Poirier L, Lamothe F, et al. Predictors of asymptomatic Clostridium difficile colonization on hospital admission. Am J Infect Control. 2015;43(3):248-253.
doi pubmed - Loo VG, Bourgault AM, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365(18):1693-1703.
doi pubmed - Kamboj M, Brite J, McMillen T, Robilotti E, Herrera A, Sepkowitz K, Babady NE. Potential of real-time PCR threshold cycle (CT) to predict presence of free toxin and clinically relevant C. difficile infection (CDI) in patients with cancer. J Infect. 2018;76(4):369-375.
doi pubmed - Britton RA, Young VB. Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol. 2012;20(7):313-319.
doi pubmed - Larcombe S, Hutton ML, Lyras D. Involvement of bacteria other than clostridium difficile in antibiotic-associated diarrhoea. Trends Microbiol. 2016;24(6):463-476.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Clinical Medicine Research is published by Elmer Press Inc.


